Alfalfa stress response gene MsNAC3 and application thereof
An alfalfa and gene technology, applied in the field of genetic engineering, can solve the problems of slow growth and low yield, and achieve the effects of improving cold resistance and stress resistance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Acquisition of alfalfa stress response gene MsNAC3
[0038] The present invention designs homologous primers based on the alfalfa MsNAC1 (JN099384) sequence:
[0039] Upstream primer: 5'-ATGGAAAGAACTCACTTCAATATC-3' (SEQ ID NO.1)
[0040] Downstream primer: 5'-GAAAAGGTTTTGGGCAAGTAC-3' (SEQ ID NO.2)
[0041] The alfalfa seedlings were placed at 4°C for cold stress 4h before the experiment. Take young alfalfa leaves and extract total RNA. TaKaRa RNAiso Plus is used for total RNA extraction. Then, alfalfa total RNA is used as a template and Oligod (T) is used as a primer for reverse transcription reaction. The reaction system is as follows:
[0042]
[0043] The reaction procedure is: 42°C for 60 minutes, 70°C for 15 minutes, and the reaction product is stored at -20°C for later use.
[0044] Using the above cDNA as a template for PCR, the reaction system is as follows:
[0045]
[0046]
[0047] The PCR reaction program is 94℃3min, (94℃10s, 55℃20s, 7...
Embodiment 2
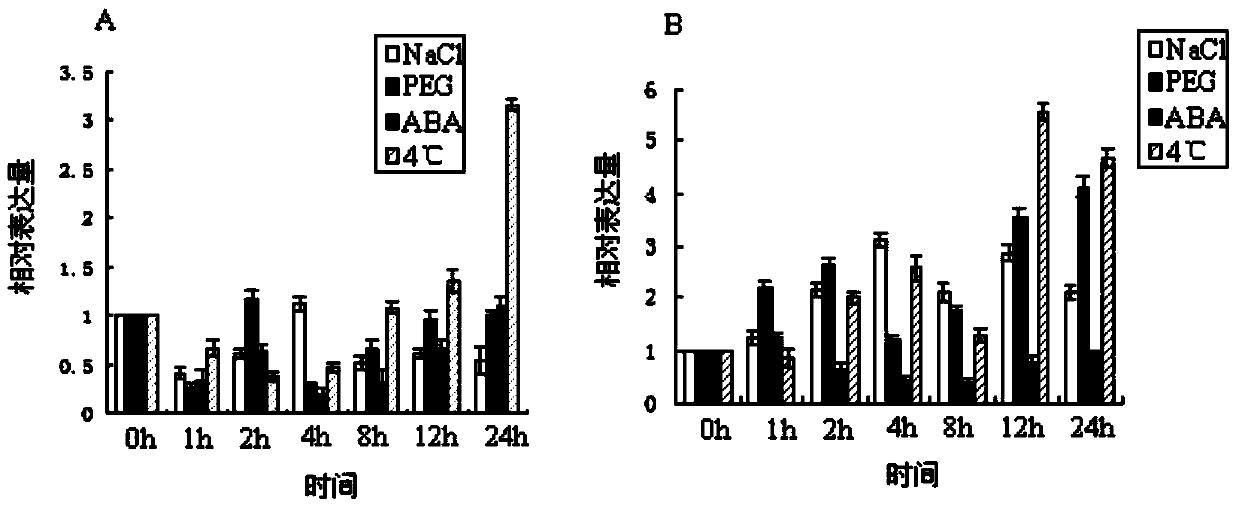
[0050] Example 2 Real-time fluorescent quantitative PCR to detect the expression characteristics of MsNAC3 gene in alfalfa under stress conditions
[0051] The plump alfalfa seeds were selected and sown on MS solid medium and cultured in a light incubator with a light cycle of 16h / d. Then, the two-week-old seedlings were transferred to each containing 250mmol·L -1 NaCl, 10%PEG6000, 100μmol·L -1 Abscisic acid (ABA) MS liquid medium was subjected to stress treatment, and chilling treatment was carried out in a refrigerator at 4°C. The treatment time of NaCl, PEG6000, ABA and 4℃ was 1, 2, 4, 8, 12 and 24h. The untreated ones were used as controls. After the treatments were completed, the total RNA of the leaves and roots of each treatment was extracted and stored in -70℃ refrigerator for use.
[0052] Design fluorescent quantitative expression primers based on the full-length cDNA sequence of MsNAC3:
[0053] Upstream primer: 5'-TGTTGATGCCACTCTGAACAC-3' (SEQ ID NO.3)
[0054] Downstrea...
Embodiment 3
[0060] Example 3 Subcellular localization of MsNAC3 gene
[0061] In order to further study the mechanism of MsNAC3 protein, a cell location detection vector fused with reporter gene EGFP was constructed, combined with Agrobacterium-mediated transient expression technology and laser confocal detection technology, to clarify the subcellular location of MsNAC3 protein.
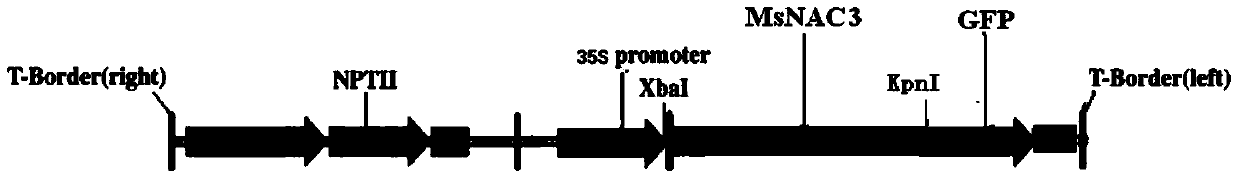
[0062] Design primers based on the sequence of the complete open reading frame of MsNAC3:
[0063] Upstream primer: 5'-TCTAGAATGGAAAGAACTCACTTCAATATC-3' (SEQ ID NO.7)
[0064] Downstream primer: 5'-GGTACCGAAAAGGTTTTGGGCAAGTAC-3' (SEQ ID NO.8)
[0065] Using the cDNA reversed from the total RNA of alfalfa leaf as a template, a PCR reaction was performed. The amplified product was recovered and connected to the pMD18-T vector for sequencing. The plasmid was extracted from a single positive colony that was sequenced correctly, and the plasmid was digested with restriction enzymes XbaI and KpnI. The target fragment was reco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com