High throughput screening model for hepatitis C virus (HCV) resistant drugs and application thereof
An anti-hepatitis C, high-throughput technology, applied in genetic engineering, cell biology, and medical fields, can solve problems such as unsatisfactory, human factors and large errors, difficult to achieve high-throughput or automated applications, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Example 1 Anti-HCV drug high-throughput screening model and construction method thereof
[0080] 1. Construction of reporter system plasmid
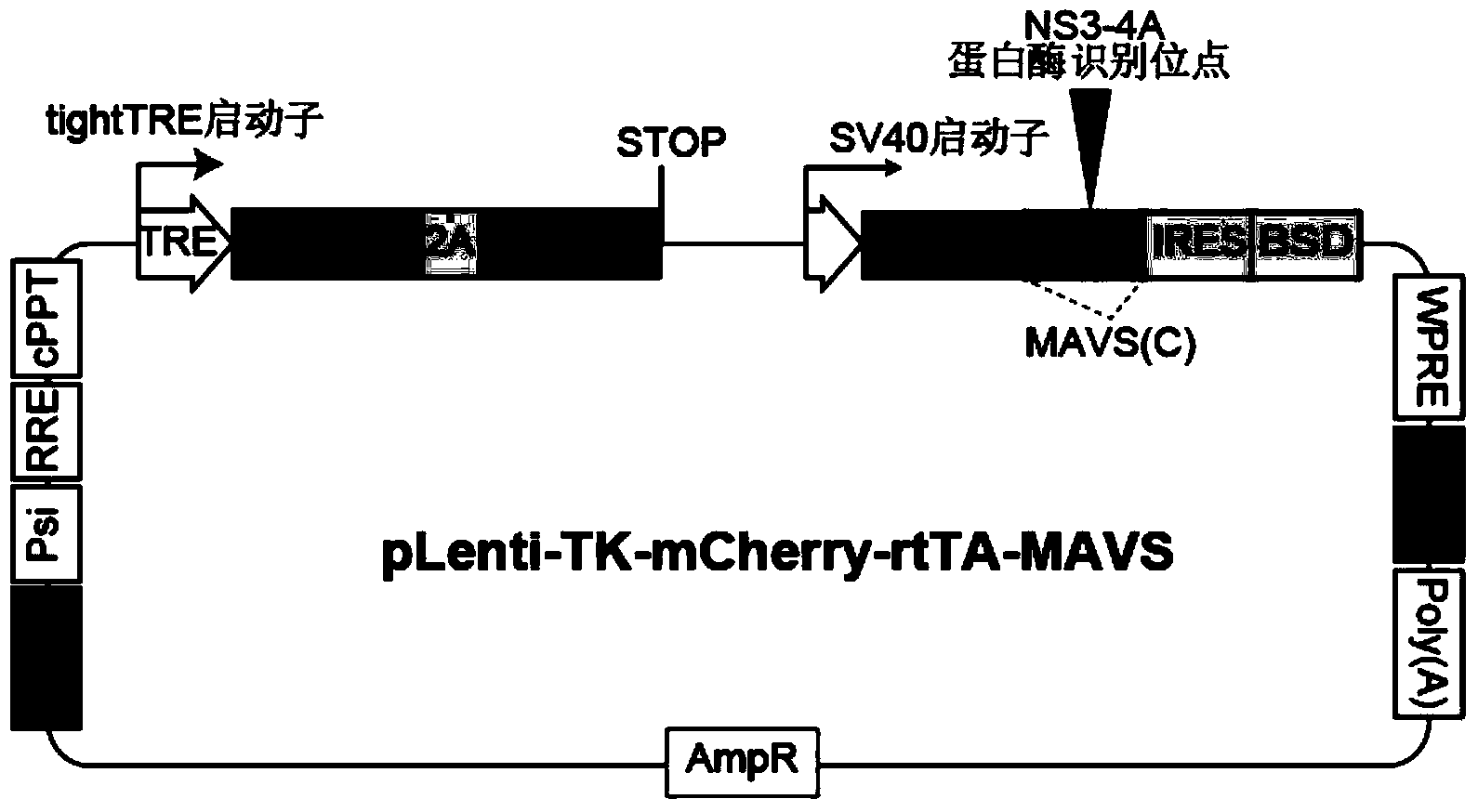
[0081] Using the plasmid pLentiCMVMCSSBsd as the backbone, the plasmid map is as follows Figure 12 As shown, the full sequence of the plasmid is shown in Seq ID No.9.
[0082] Cloning is a two-step process:
[0083] Step 1: Delta-tk, 2A, and mCherry reporter gene modules correspond to primers 1-6 in Table 1, respectively, and are amplified by PCR. The gene templates are stored by Wei Wensheng Laboratory, School of Life Sciences, Peking University. Then the corresponding restriction endonucleases were used to carry out enzyme digestion, and sequentially loaded into the multiple cloning site behind the tight-TRE promoter of the pEN_TTmcs plasmid. Thus, the complete sequence from tight-TRE to mCherry (including stop codon) was obtained.
[0084] Then, the CMV promoter that comes with the pLentiCMVMCSSBsd plasmid was excised with...
Embodiment 2
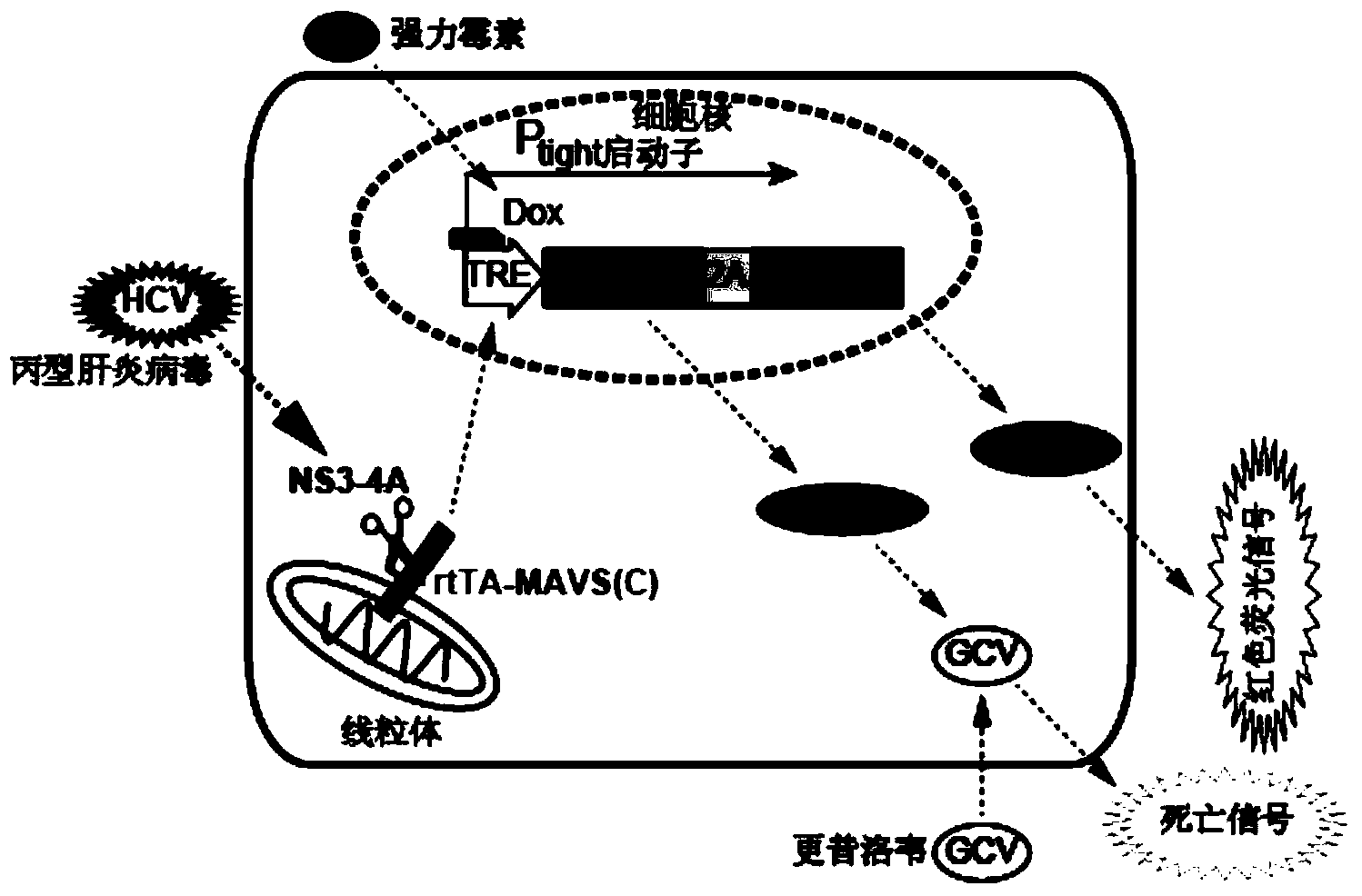
[0097] Example 2 NS3 / 4A specific inhibitor VX-950 can significantly inhibit the fluorescent signal of the system
[0098] This example was developed based on the physiological activity of HCV NS3 / 4A protease. In order to verify the rigor and sensitivity of the reporter system constructed in Example 1, and whether it can be applied to compound screening, the following experiments were designed:
[0099] The HCV host cell Huh7.5 containing the reporter system was mixed with 10 per well 5 The cells were spread evenly on a 12-well plate. Cultured in DMEM medium at 37°C in an incubator with 5% carbon dioxide concentration.
[0100] The DMEM medium is DMEM (Life Technologies), 10% FBS (Thermo Scientific), 1×NEAA (Life Technologies) and contains 1× double antibody (penicillin-streptomycin).
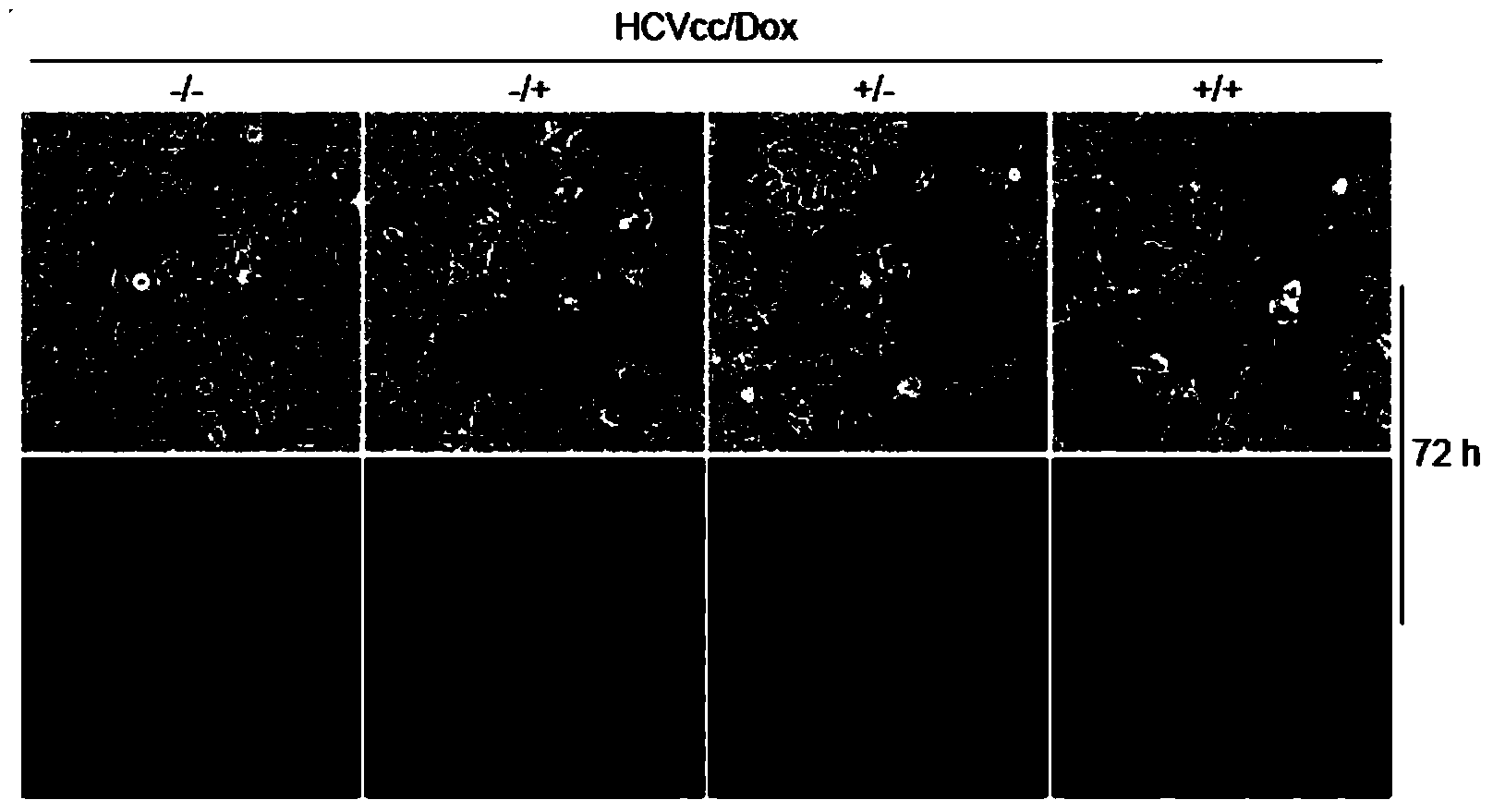
[0101] The next day, use doxycycline (2 μg / ml) and HCVcc (3.6×10 4 FFU / ml) of DMEM to renew the medium in each well.
[0102] At the same time, with DMSO as the control group, different volu...
Embodiment 3
[0104] Example 3 Using Flow Cytometry (FACS) to Realize High Throughput Analysis
[0105] In order to further verify the sensitivity of the reporter system of the present invention to HCVcc infection, and whether FACS can be successfully used for high-throughput analysis, the following experiments were designed:
[0106] 1. Prepare cells and set up analysis samples
[0107] 2×10 per well 5 Density of cells The monoclonal Huh7.5 cells with the best performance selected in Example 1 were spread in turn on a 6-well plate, with a total of 14 wells, including a control group and 13 experimental groups.
[0108] On the next day, referring to the method of using the fluorescent signal in Example 2, the medium was renewed with DMEM containing doxycycline, 2 ml per well. Add 0.6ml of HCVcc virus stock solution and 0.4ml of DMEM medium (and supplement 1μl of doxycycline stock solution with a concentration of 2mg / ml) at the highest concentration (well 14), a total of 3ml, and the calcu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com