Recombinant pichia pastoris bacterial strain for co-expressing inulin excision enzyme and incision enzyme as well as construction method and application of bacterial strain
A technology of exo-inulinase and Pichia pastoris, which is applied in the field of genetic engineering and fermentation engineering, can solve the problems of unsuitable industrial application, difficulty in large-scale cultivation, low enzyme activity, etc., achieve good genetic stability, and is fully feasible The effect of improving the efficiency and enzyme digestion efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
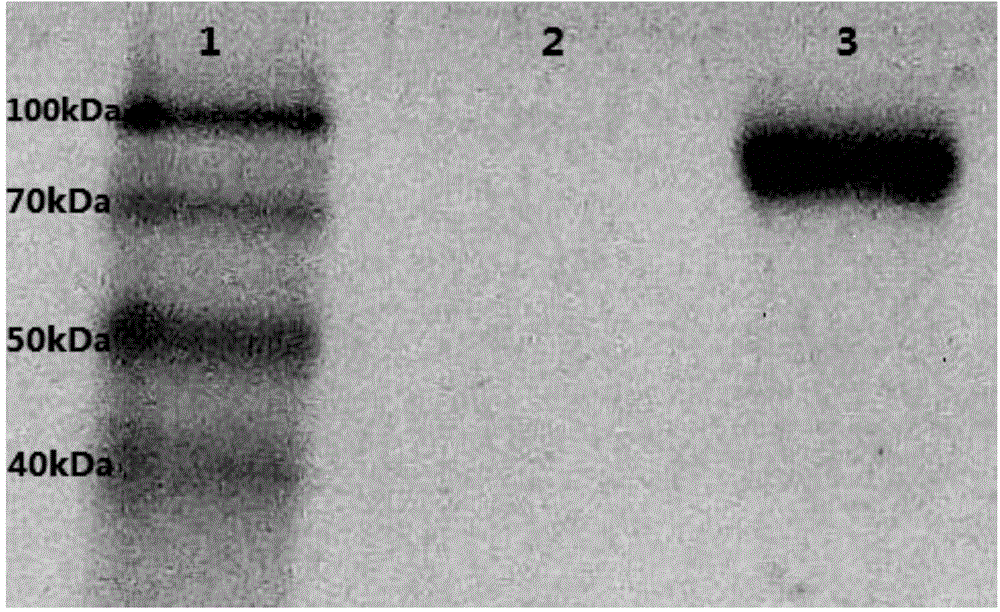
[0037] Example 1 Construction of recombinant Pichia pastoris GS115-INU1 expressing exo-inulinase INU1
[0038] 1. Construction of recombinant plasmid pPIC9-INU1
[0039] 1.1 Genome extraction
[0040] The genome of Kluyveromyces marxianus CGMCC2.1440 was extracted with a yeast genomic DNA extraction kit (purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd.). For specific preparation methods, refer to the operation manual of the kit.
[0041] 1.2 Cloning of gene INU1
[0042] Use the genome extracted in 1.1 as a template for the PCR reaction, and design a pair of primers based on the known exinulinase gene sequence on NCBI:
[0043] INU1-XhoI upstream:
[0044] GCCCG CTCGAG AAAAGAGAGGCTGAAGCTAGAGATGGTGACAGCAAGGCC
[0045] (The underlined part is the XhoI restriction site);
[0046] Downstream of INU1-AvrII:
[0047] GCCCG CCTAGG ATGGTGGTGATGGTGGTGAACGAACGTTACCCAATTTAACG
[0048] (the underlined part is the AvrII restriction site),
[0049] A PCR reactio...
Embodiment 2
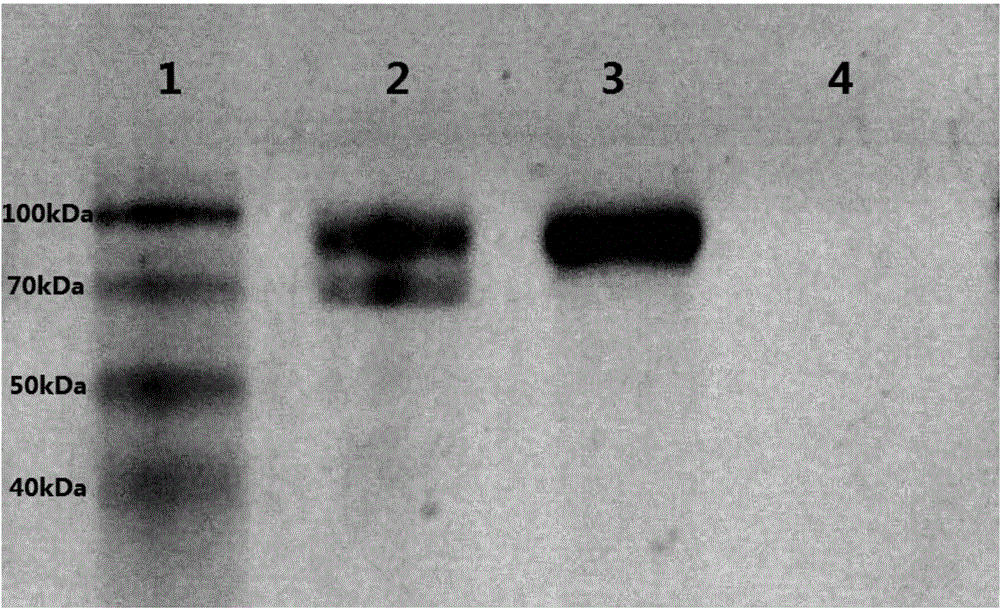
[0084] Example 2 Construction of recombinant Pichia pastoris GS115-INU1-INU2 expressing exinulinase INU1
[0085] 1. Construction of recombinant plasmid pPIC9K-INU2
[0086] 1.1 Genome extraction
[0087]The genome of Aspergillus ficuum CGMCC3.4322 was extracted with Biospin Fungus Genomic DNA Extraction Kit (Bioer Technology co., Ltd.). For the specific preparation method, refer to the operation manual of the kit.
[0088] 1.2 Cloning of gene INU2
[0089] Use the genome extracted in 1.1 as a template for the PCR reaction, and design a pair of primers based on the known endinulinase gene sequence on NCBI:
[0090] INU2-EcoR I upstream:
[0091] ACGC GAATTC CAGTCTAATGATTACCGTCCTT (the underlined part is the EcoR I restriction site);
[0092] Downstream of INU2-Not I:
[0093] ATA GCGGCCGC TCATTCAAGTGA AACACTCC (the underlined part is the Not I restriction site),
[0094] A PCR reaction was performed to clone INU2. PCR amplification conditions: pre-denaturation at 94...
Embodiment 3
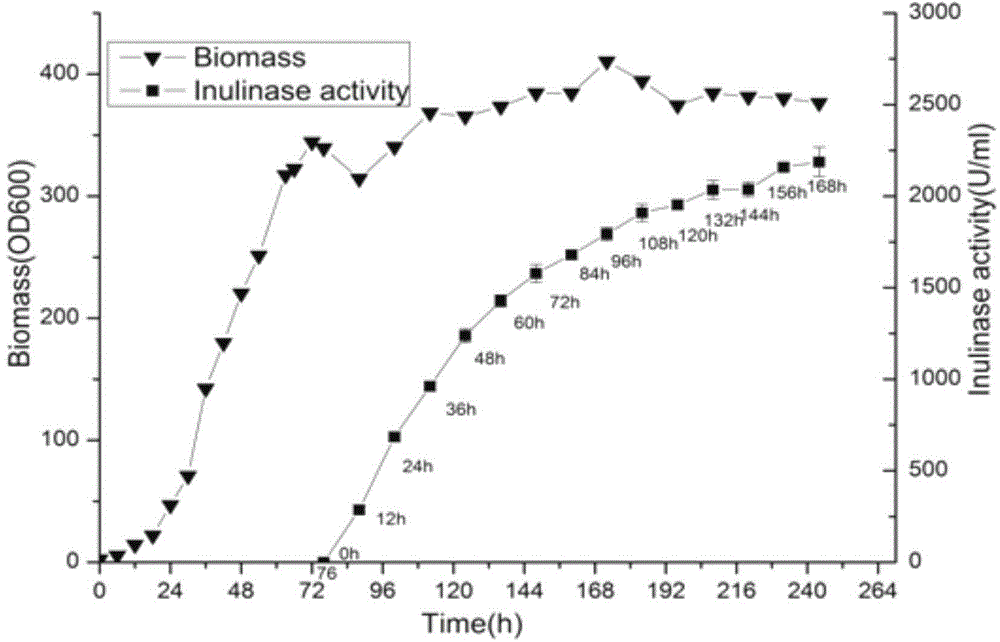
[0127] Embodiment 3 Utilize the method for expressing inulinase in large quantities by high-density fermentation of Pichia pastoris (Pichia pastoris) GS115-INU1-INU2 strain of the present invention
[0128] The main instruments used in this method are:
[0129] Fermenter Labfors5 (3.6L): purchased from Iverson Biotechnology Co., Ltd.
[0130] The specific plan is as follows:
[0131] Inoculate the transformant GS115-INU1-INU2 into 100ml of YPD medium, and shake the flask to culture to bacterial concentration OD 600 The value is 4-6, and the inoculation amount of 10% by volume is received in a 3L fermenter equipped with 1.2LBSM basic salt medium, and fed-batch culture is carried out. During the fermentation process, ammonia water is added to adjust the pH value, the ventilation volume is maintained at 1-3vvm, and the rotation speed is 400-900r / min.
[0132] Bacteria growth stage: temperature is set at 30°C, pH is 4.5, and dissolved oxygen is kept above 10%. After 22-24 hours...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com