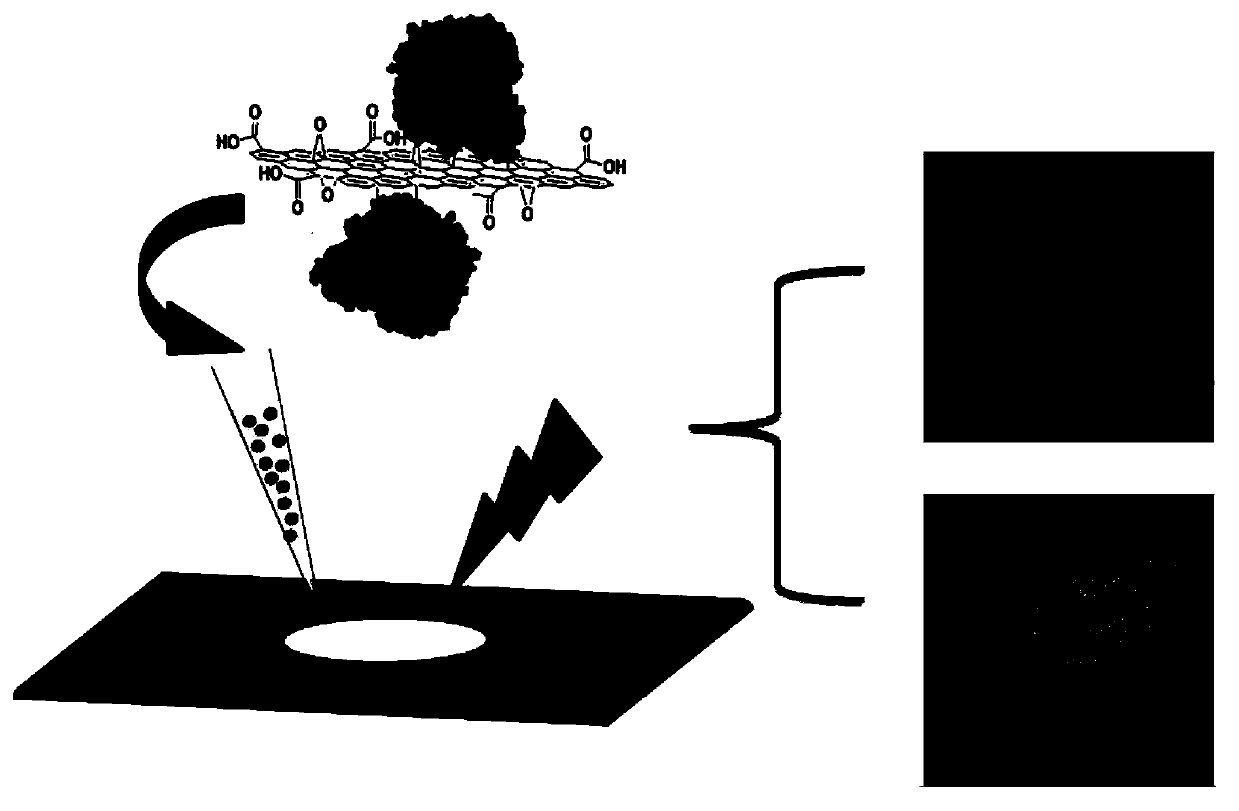
A method for protein identification and imaging mass spectrometry based on efficient in situ enzymatic hydrolysis of immobilized enzyme
An immobilized enzyme, mass spectrometry imaging technology, applied in the field of biochemical analysis, can solve problems such as affecting the detection of low-abundance proteins, ineffective enzyme digestion, low protein content, etc., and achieves good aqueous solution stability, simple operation and simple steps. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Investigation and Experiment of Graphene Oxide Material Interfering with Mass Spectrometry Detection Signal
[0050] A bovine serum albumin enzymatic peptide solution with a concentration of 5 ng / μL and its mixed solution with the graphene oxide material dispersion (prepared at a ratio of 1:1) were prepared respectively. Take 1 μL of the samples and spot them on the MALDI target plate, and then spot an equal volume of CHCA matrix solution (0.1% trifluoroacetic acid 50% acetonitrile aqueous solution) after drying. MALDI-TOF / MS analysis was carried out after drying and crystallization, the results are as follows figure 2 shown.
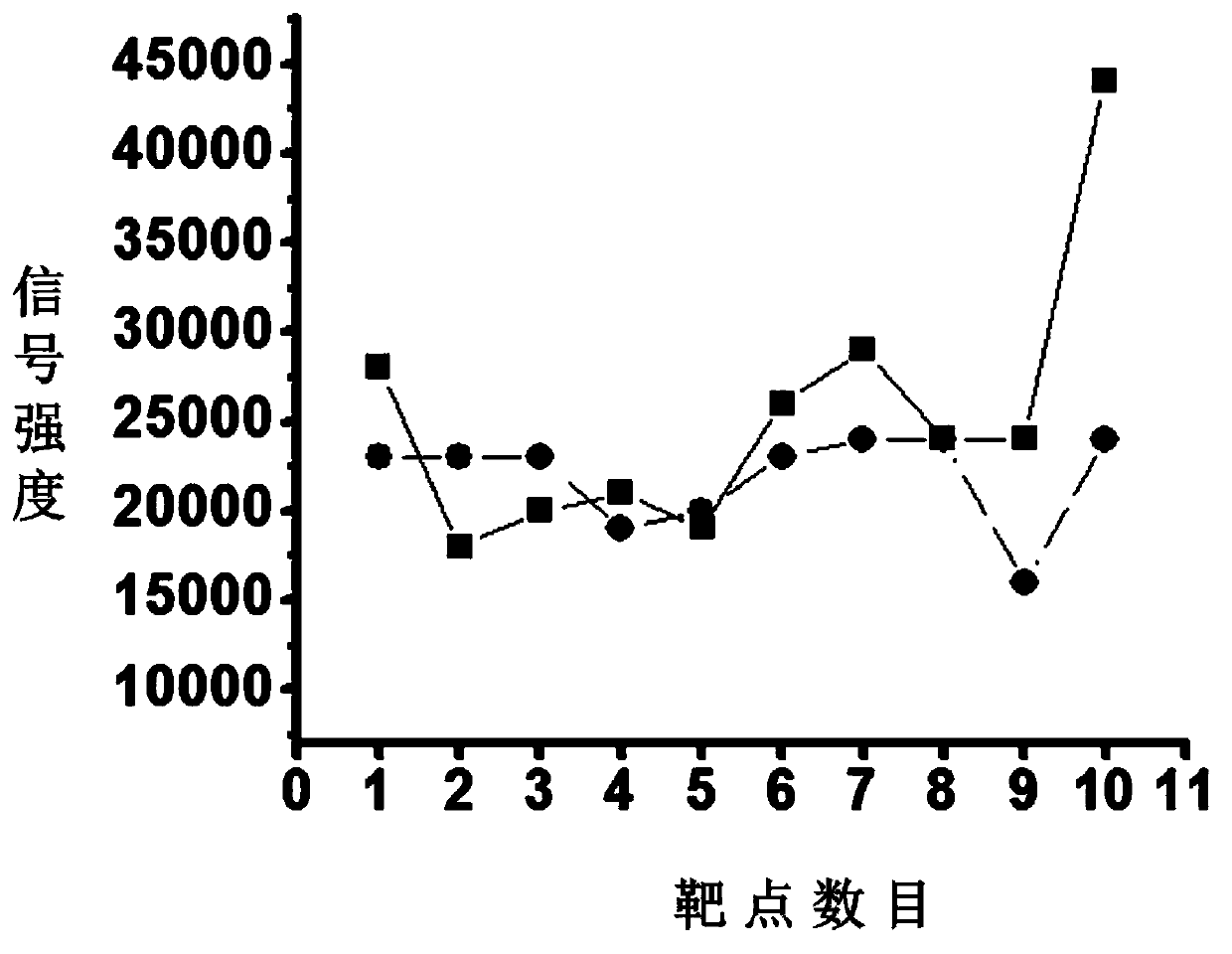
[0051] A bovine serum albumin enzymatic peptide solution with a concentration of 10 ng / μL and its mixed solution with the graphene oxide material dispersion (prepared at a ratio of 1:1) were prepared respectively. Take 1 μL of the samples and spot them on the MALDI target plate, and spot the samples on ten target spots continuously. After dryin...
Embodiment 2
[0053] Experiment on the Adsorption Ability of Graphene Oxide Material to Trypsin
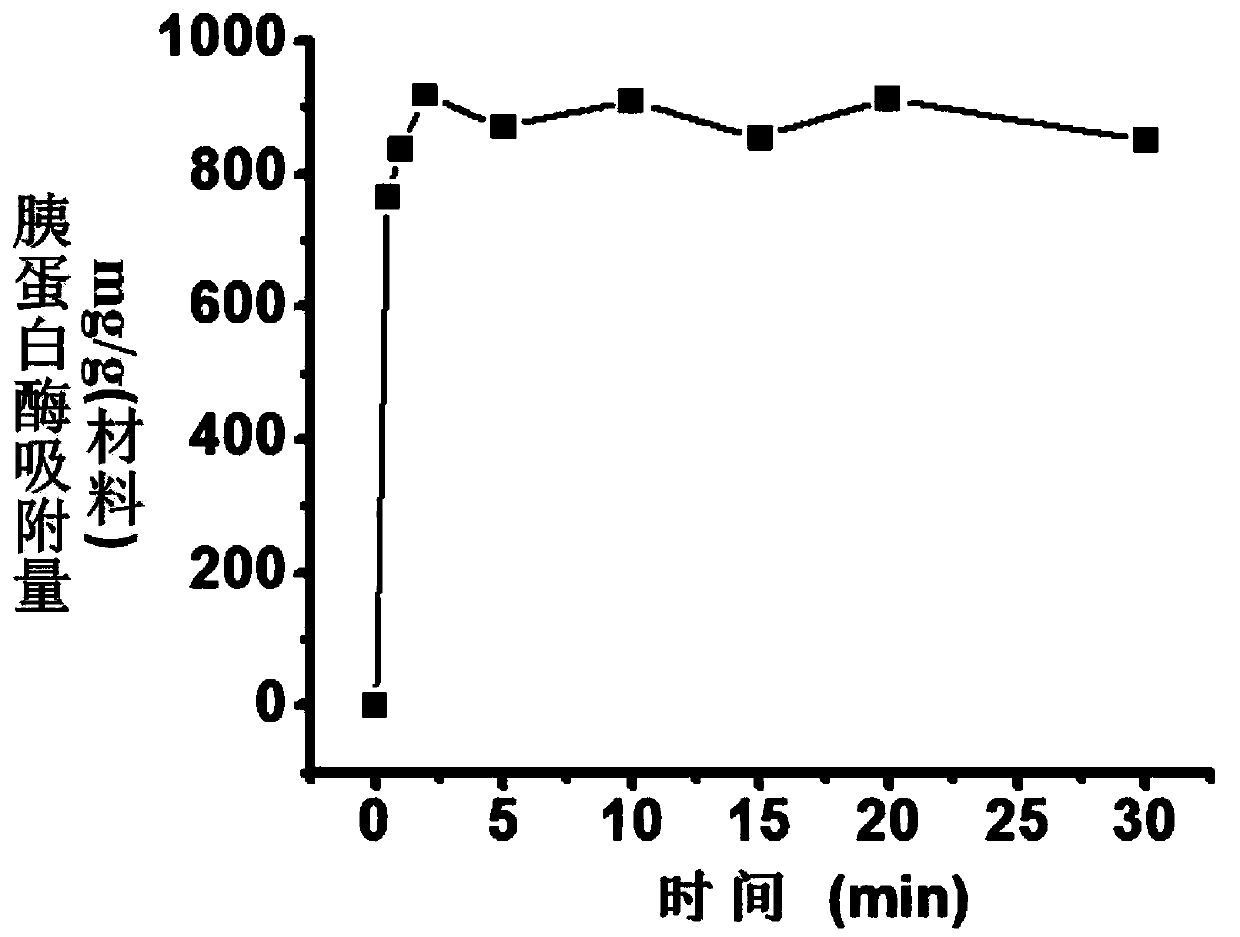
[0054] Prepare graphene oxide material dispersion and trypsin solution with a final concentration of 400 ng / μL in 20 mM phosphate buffer solution (pH5), mix and incubate in equal proportions for a certain period of time, then centrifuge for 5 minutes to take the supernatant. The concentration of unfixed trypsin in the supernatant was determined using the BCA protein concentration assay. According to the concentration of trypsinogen and the concentration of unfixed trypsin in the supernatant, the adsorption amount of trypsin by the graphene oxide material was obtained. The result is as Figure 4 shown.
[0055] The above experiments were repeated under different pH conditions (pH 4-10). The result is as Figure 5 shown.
Embodiment 3
[0057] Study on the effect of immobilized enzyme and free enzyme on enzymolysis of standard protein target
[0058] After fixation of trypsin (pH 5) as described in Example 2, it was diluted 10-fold in 25 mM ammonium bicarbonate buffer.
[0059] Prepare an aqueous solution of α-casein (10ng / μL), take 1μL of the above solution and spot on the MALDI target plate, after drying, spot the free enzyme or immobilized enzyme diluted with 25mM ammonium bicarbonate on the corresponding target , and placed in an incubator at 37 degrees Celsius for enzymatic hydrolysis. After the enzymatic hydrolysis, 1 μ LDHB matrix solution (0.1% trifluoroacetic acid 50% acetonitrile aqueous solution) was spotted on the MALDI target plate, dried and crystallized for MALDI-TOF / MS analysis, the results are as follows Figure 7 shown.
[0060] Bovine serum albumin and horse heart blood myoglobin (10ng / μL) were enzymatically hydrolyzed by the above method and then searched by mascot database search softwa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Slice thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com