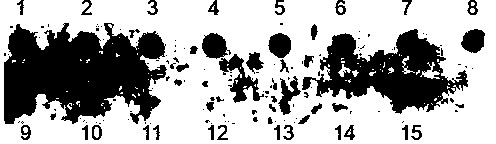Method for quickly and sensitively detecting micromolecule RNA
A sensitive detection, small molecule technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as bands cannot be displayed, background interference, and probes cannot be removed, so as to shorten the detection time and improve The effect of detection sensitivity and detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] In this example, the detection of Osa-miR156 in rice leaves, let-7-a in mouse hepatocytes, and chicken blood was taken as an example.
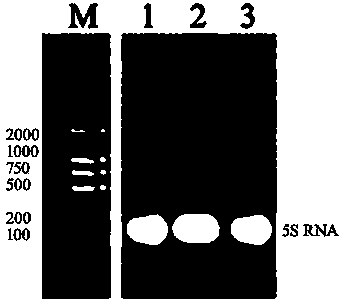
[0038]The flow chart of the whole detection method is as follows: figure 1 shown.
[0039] In the process of realizing the object of the present invention, the following steps are usually included:
[0040] 1. Material selection: fresh animal and plant tissues, blood, or fresh -70°C frozen samples.
[0041] 2. Reagent configuration:
[0042] 1) DEPC treated water RNase free water: deionized water is treated by Millpore water processor, add 1 / 1000 DEPC water for treatment, sterilize under high temperature and high pressure for 30 minutes, and cool to room temperature;
[0043] 2) TBE: Tris: 108g; Na 2 EDTA·2H 2 O: 7.44g; boric acid: 55g, add 800ml of deionized water, stir to dissolve, add deionized water to 1L, store at room temperature;
[0044] 3) 10% APS: 1g ammonium persulfate, dissolved in 10ml double distilled water;
[0...
Embodiment 2
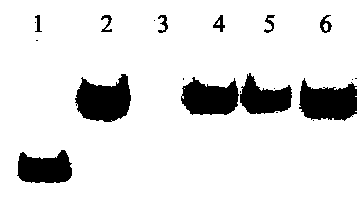
[0083] Detection sensitivity test
[0084] This embodiment is based on the method of Example 1. During liquid phase hybridization, Osa-miR156 in rice leaves with different concentrations was used for liquid phase hybridization and solid phase detection, and the detection sensitivity test was carried out. The results are as follows: Figure 4 as shown, Figure 4The concentrations of Osa-miR156 in bands 1-15 are 1. 10 fmol; 2. 5 fmol; 3. 2.5 fmol; 4. 1 fmol; 5. 0.5 fmol; 6. 0.25 fmol; 7. 0.1 fmol; 8. 0.05 fmol; 9. 0.025 fmol; 10. 0.01 fmol; 11. 0.005 fmol; 12. 0.0025 fmol; 13. 1 amol;
[0085] from Figure 4 It can be seen that this method can detect at least 0.005 fmol of miRNA (5aM=35fg=0.035pg), that is, band 11 can be clearly identified, and the sensitivity of this method is 10 times higher than that of the previous small RNA detection method.
Embodiment 3
[0087] Small molecule RNA quantitative detection test
[0088] In this example, the quantification of miR156 in rice is taken as an example; the DNA form miD156 of miR156 with a series of concentration gradients (0.8-31.2 fmol) is hybridized with 0.1 pmol of biotin-labeled probe miD156*, and after color development, Chemi Doc The XRS detection system was used for relative quantitative analysis. The concentration of miD156 used for hybridization was used as the X axis, and the chromogenic concentration was used as the Y axis to establish a coordinate curve. The results showed a good linear relationship between the two. . Band 7 is the hybridization of 1ug rice miRNA with 0.1pmol miD156* probe, the result is in the regression equation,
[0089] The result is as Figure 5 as shown, Figure 5 In , the miR156 concentrations of bands 1-7 were 1: 0.8fmol; 2: 1.9fmol; 3: 3.9fmol; 4: 7.8fmol; 5: 15.6fmol; 6: 31.2fmol.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com