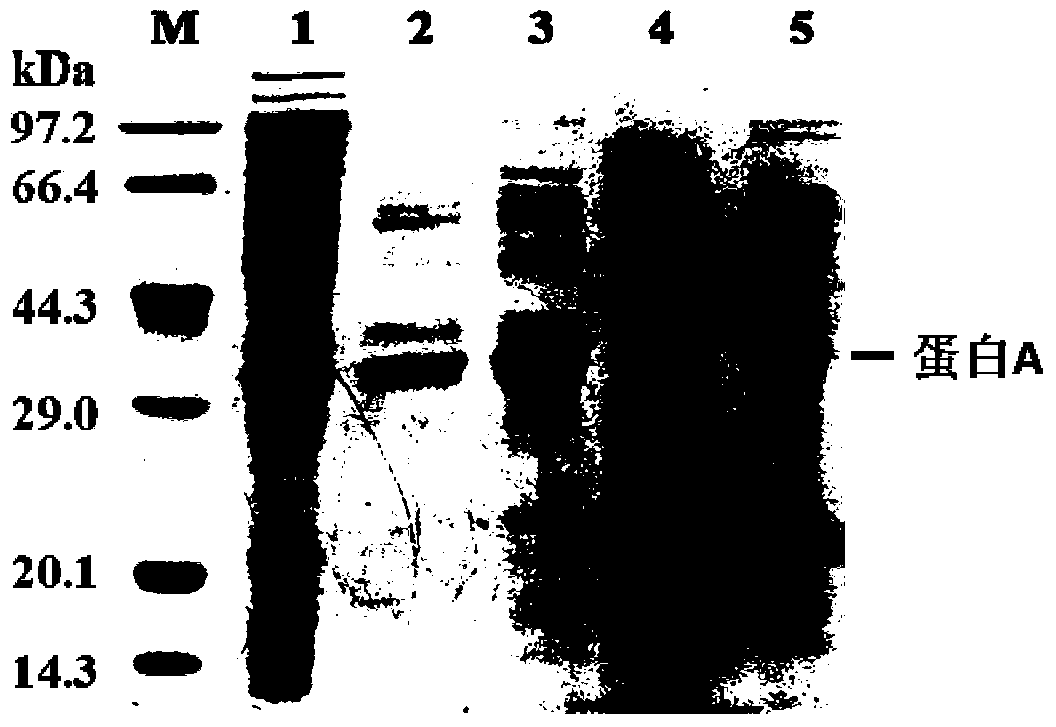Fixed-point immobilization method for recombinant protein A affinity ligands
A fixed-point and affinity technology, applied in chemical instruments and methods, separation methods, carrier binding/immobilization of peptides, etc., can solve the problems of destroying disulfide bonds, affecting the effect of fixed-point immobilization, and high production cost of protein ligands. The effect of improving adsorption efficiency, reducing selling price and reducing production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1. Preparation of protein A modified by site-directed formhylation
[0030] 1. Vector construction and transformation screening:
[0031] Protein A gene sequence: The signal peptide and membrane binding domain are removed from the full-length gene of protein A, and the gene sequences of five antibody Fc fragment binding domains A, B, C, D, and E are used as the gene sequence of protein A. The full length is 888bp, as shown in SEQ ID NO.2.
[0032] Through two rounds of PCR amplification, insert the specific sequence recognized by FGE shown in SEQ ID NO.1 into the 5' end of the protein A gene sequence, the specific method is:
[0033] (1) The names and sequences of the primers used are shown in Table 1, and the underlined part is the enzyme cutting site.
[0034] Table 1. Primer names and their sequences
[0035]
[0036] (2)PCR
[0037] The first round of PCR: the protein A gene sequence was ligated into the pET23a plasmid through the Nde Ⅰ and BamH Ⅰ res...
Embodiment 2
[0046] Example 2. Preparation of surface hydrazide functionalized agarose gel microspheres:
[0047] The preparation method of the solid phase chromatographic matrix with active hydrazide groups includes the activation of epichlorohydrin, the addition of diamino reagents for epoxy ring opening, further addition of dialdehyde reagents to form a chromatographic matrix with active aldehyde groups, and finally the use of bisamino reagents to form a chromatographic matrix with active aldehyde groups. The hydrazide reagent reacts with the aldehyde group on the chromatographic matrix to form a solid phase chromatographic matrix with active hydrazide groups. Wherein the diaminopropyl imine is preferred as the diamino reagent, glutaraldehyde is preferred as the dialdehyde reagent, and adipate dihydrazide is preferred as the dihydrazide reagent. The specific method is as follows:
[0048] (1) Epichlorohydrin activation: Take 10mL of Sepharose CL-4B, wash it with single distilled water ...
Embodiment 3
[0052] Example 3. Direct site-specific immobilization of aldehyde protein A
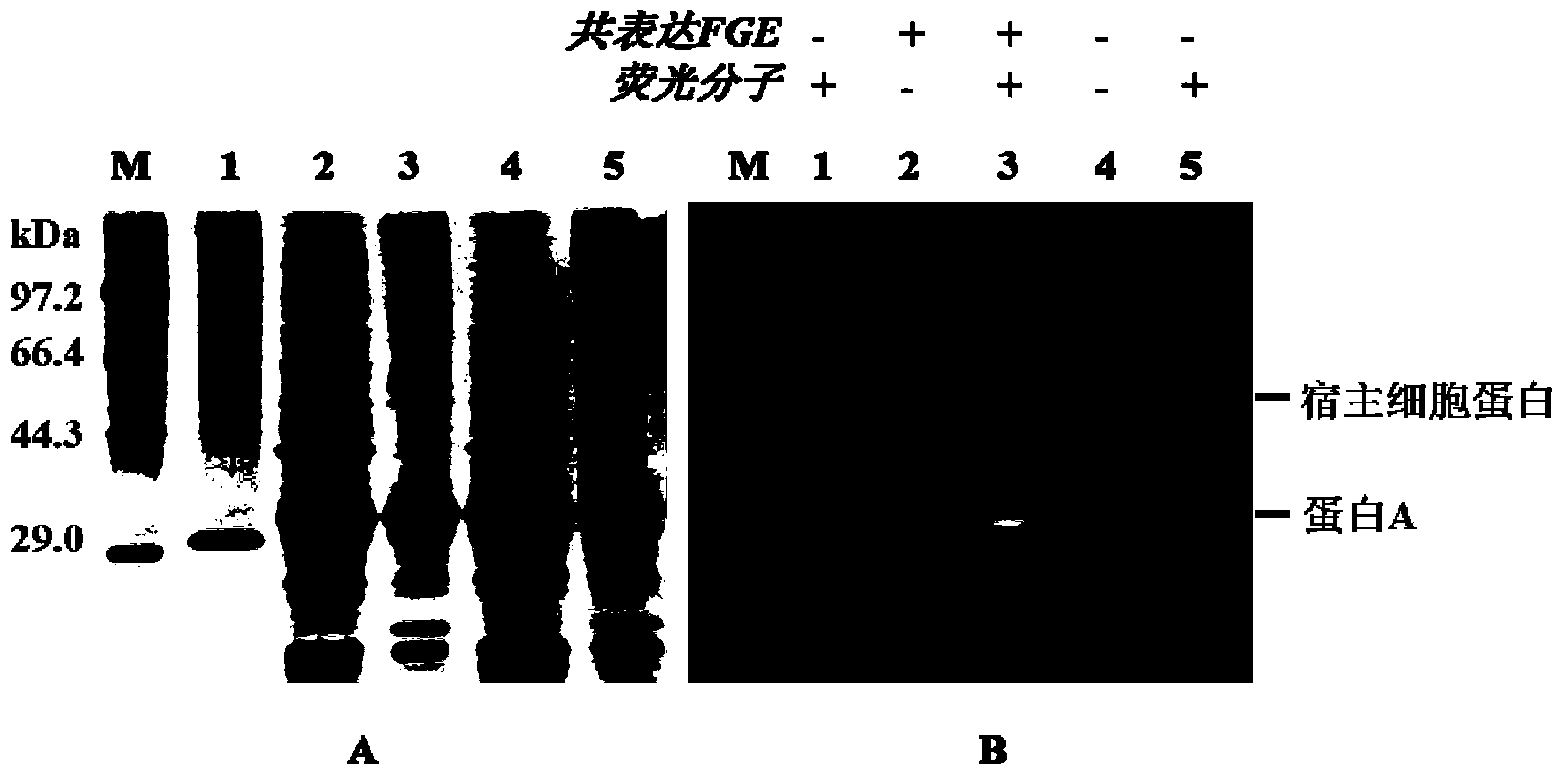
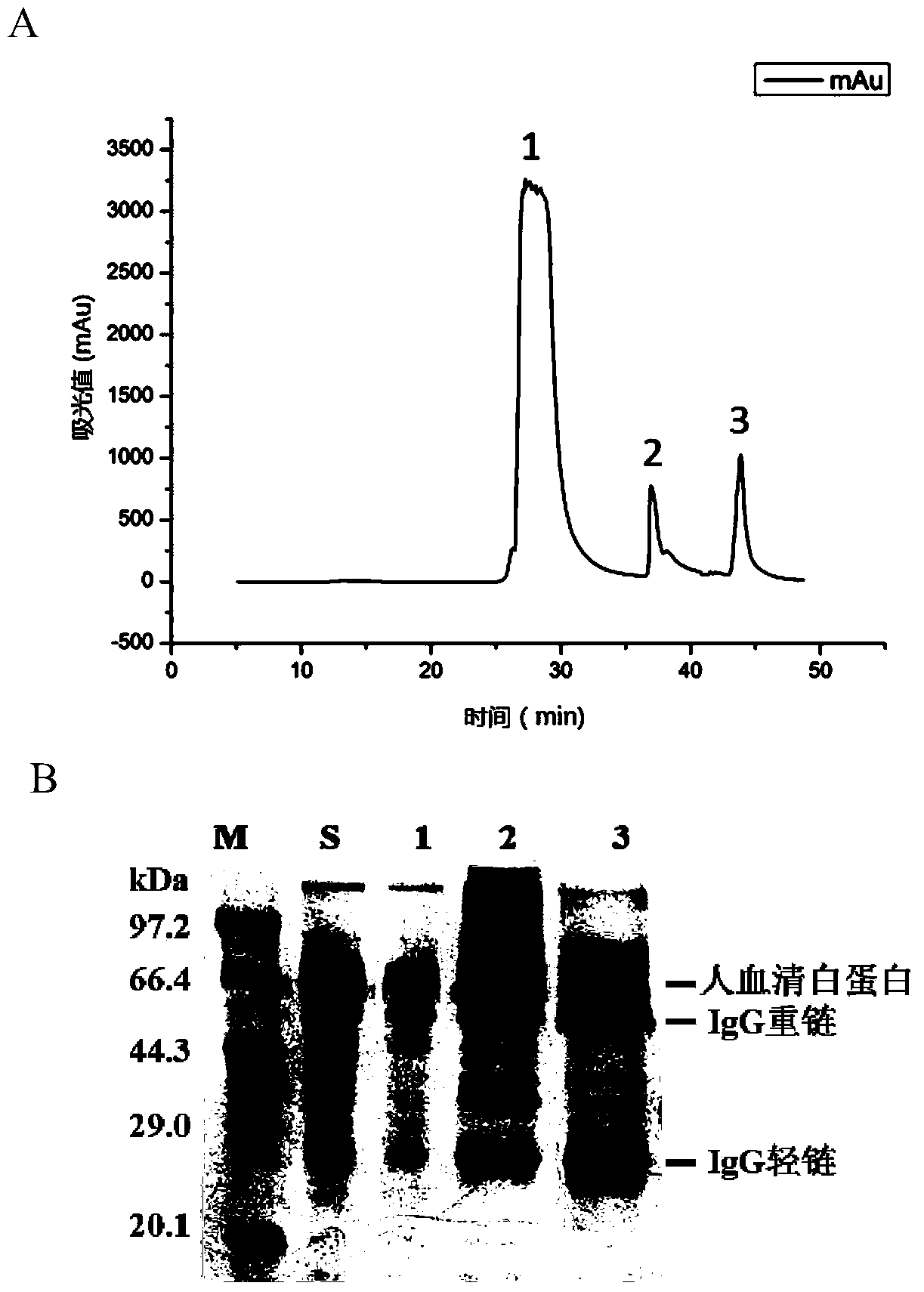
[0053] Take 4mL of the hydrazide functionalized agarose gel in Example 2, divide it into four equal parts, each 1mL, add 1mL of the cell crushing supernatant in Example 1 respectively, in order to investigate the reaction of different catalysts on hydrazide and aldehyde groups 100mM aniline, 10mM p-phenylenediamine, and 5mM 3,5-diaminobenzoic acid were added to the reaction system as catalysts, and PBS was added as a control. After reacting at 37°C for 12 hours, the solid-phase matrix was collected by centrifugation, and the solid-phase matrix was collected with 1M The solid-phase matrix was washed 3 times with PBS of NaCl to wash away non-specifically bound proteins. The chromatographic matrix of site-fixed protein A was obtained and stored in PBS solution containing 0.02% sodium azide at 4°C. Further, 200mM hydroxylamine hydrochloride was used to cut off the hydrazide and aldehyde group under the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com