Thioether monooxygenase from pseudomonas monteilii as well as synthesis and application thereof
A thioether monooxygenase and protein technology, which is applied in the fields of biocatalysis, biosynthesis and molecular biology, can solve the problems of green industrial production, low catalytic efficiency, and lack of highly enantioselective thioether monooxygenase And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1. Acquisition of monooxygenase-encoding genes
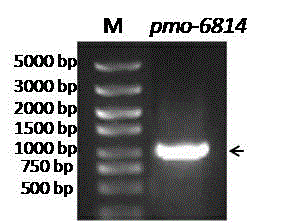
[0031] By analyzing the genome sequence of Pseudomonas strain, a 972 bp monooxygenase sequence was determined, a pair of specific primers were designed to amplify by PCR technology, and finally the full-length coding frame sequence of the gene was obtained. The specific method is as follows: using the extracted genomic DNA as a template, use primers F: 5’-actcGGATCCatgcagaccgttgaacacgaat-3’ and R: 5’-accgAAGCTT
[0032] gtaggctttgtcccagct-3' for PCR amplification. The PCR reaction system is as follows: 12.5 ul of 2×TaqPCR Master Mix, 1 ul of genomic DNA, 0.5 ul of upstream and downstream primers, and 10.5 ul of ddH2O. PCR reaction conditions: 95°C for 10 min; cycle of 98°C for 10 s, 56°C for 30 s, 72°C for 90 s; extension at 72°C for 10 min, 30 cycles. The PCR product was electrophoresed on a 1% agarose gel to verify whether a fragment conforming to the size of the target gene was obtained.
Embodiment 2
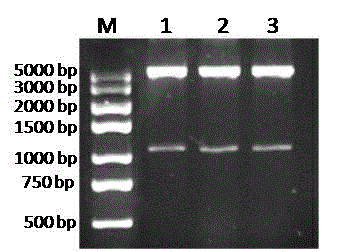
[0033] Example 2. Monooxygenase gene expression vector construction
[0034] The monooxygenase gene was constructed using the pET32a(+) vector (purchased from Clonetech) as the backbone pmo-6814 prokaryotic expression vector. The specific method is as follows: Use the DNA gel recovery kit to recover pmo-6814 The PCR product was digested with restriction endonucleases BamHI and HindIII on the PCR product and the vector pET32a(+), and the recovered target fragment and vector were digested. After ligation with T4 DNA ligase at 16 °C for 4 h, the ligation product was transformed into Escherichia coli DH5α. After overnight culture, the grown monoclonal colony was picked and shaken to extract the plasmid, and the recombinant plasmid was detected by double enzyme digestion with BamHI and HindIII. Select the positive recombinant plasmid pET32a- pmo-6814 DNA sequencing is performed to ensure the sequence is accurate. Finally, the positive recombinant plasmids with accurate sequ...
Embodiment 3
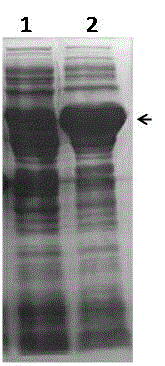
[0035] Example 3. Recombinant expression and detection of monooxygenase protein
[0036] Will be stored in glycerol containing pET32a- pmo-6814 After activation of the genetically engineered bacteria BL21(DE3) with the plasmid, pick a single colony in 3 ml of liquid LB medium containing the corresponding antibiotics, culture with shaking at 37 °C for 12 h, and transfer to a fresh medium containing 1% of the inoculum the next day. In 50 mL LB liquid medium of antibiotics, shake culture at 37°C 250 rpm with OD600 of 0.6 (about 3h), add IPTG with a final concentration of 0.5 mM, and induce culture at 25°C 160 rpm for 8h. After induction, collect the cells by centrifugation at 8,000 rpm / min for 5 min, resuspend the cells in PBS buffer, break up the cells by ultrasonic, and centrifuge at 15,000 rpm for 5 min to remove cell debris, take the supernatant and mix with 5x loading buffer, and place in SDS-PAGE electrophoresis analysis was performed after heating in a constant tempera...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com