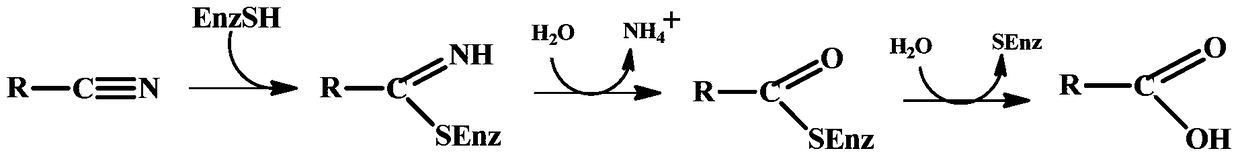
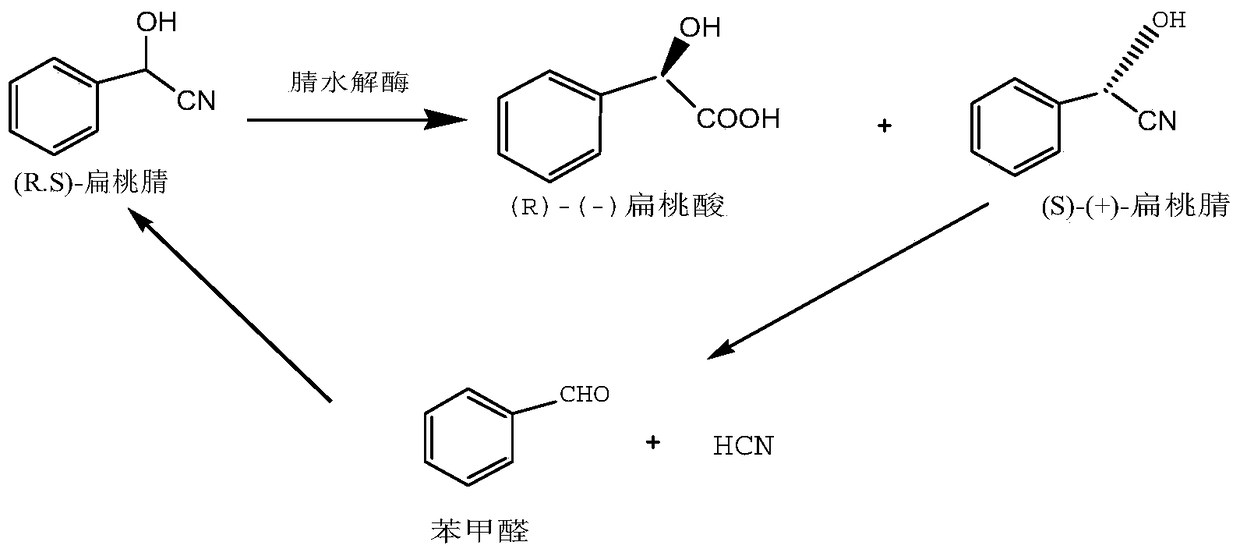
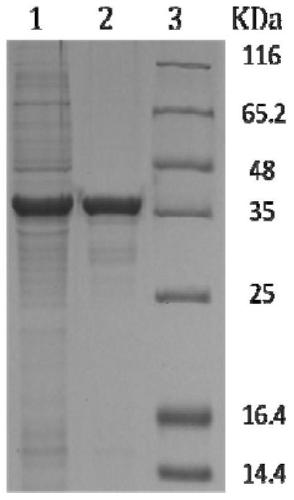
Nitrilase mutant, gene, carrier, engineering bacteria and application
A technology of nitrilase and mutants, applied in genetic engineering, hydrolase, application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Gene synthesis of parent nitrilase
[0027] Parental nitrilase gene (GeneBank: AY487562), in order to express the protein with His-tag after the gene is connected to the vector pET28b, its stop codon was excised, and the codon preference of E.coli was used as a reference pair Its sequence was optimized and NcoI was introduced at the start codon to insert glycine after methionine. The sequence of the newly designed nitrilase gene (tegi) is shown in SEQ ID NO.1 (the amino acid sequence is shown in SEQ ID NO.2). The gene synthesis was entrusted to Shanghai Xuguan Biotechnology Development Co., Ltd. After the gene synthesis Linked into the expression vector pET28b.
Embodiment 2
[0028] Embodiment 2: Construction of mutant library
[0029] Using the parental sequence of the nucleotide sequence shown in SEQ ID NO.1 obtained in Example 1 as a template, the following nucleotide sequences (Table 1) were used as primers to carry out PCR amplification respectively to obtain the nucleotide sequence shown in SEQ ID NO.2 respectively. The 190th serine of the amino acid sequence is mutated to alanine, leucine, valine, glycine, threonine, cysteine, arginine, lysine, histidine, glutamic acid mutant.
[0030] Table 1: Primers
[0031]
[0032]
[0033] The PCR reaction system is:
[0034]
[0035] The PCR program was set as follows: pre-denaturation at 95°C for 5 min; denaturation at 95°C for 15 s, annealing at 55°C for 30 s, extension at 72°C for 7 min, 30 cycles; final extension at 72°C for 10 min, and storage at 4°C.
[0036] After the PCR, 20 μL of the PCR product was taken and detected by 0.9% agarose gel electrophoresis. For the positive PCR prod...
Embodiment 3
[0040] Example 3: Screening of Mutant Library
[0041] (1) Pick the single bacterium colony cultivated in Example 2 into 50mL of LB liquid medium (containing 50ug / mL kanamycin), put it into 37°C, and cultivate it for 10h under the condition that the shaker speed is 150rpm, and then take the seeds The solution was transferred to 100mL LB liquid medium with an inoculum of 2% volume concentration, and cultured at 37°C and the shaker speed was 150rpm. When the concentration of the bacteria was OD 600 When reaching 0.6-0.8, add IPTG with a final concentration of 0.5mM. Then put into 28 DEG C, 150rpm shaker and cultivate for 8 hours, centrifuge to collect the thalline, and then wash the thalline twice with 0.85% physiological saline. Recombinant Escherichia coli bacteria confirmed as positive mutations by sequencing were stored in glycerol tubes (final concentration of glycerol was 15%).
[0042] (2) Determination of enzyme activity and definition of enzyme activity
[0043] Weig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com