Method for purifying pseudomonas aeruginosa vaccine recombinant protein Vac 9
A Pseudomonas aeruginosa purification method technology, applied in the field of biopharmaceuticals, can solve the problems of recombinant protein Vac9 purification methods and reports that have not yet been seen, and achieve good immune protection, good repeatability, and high humoral immune response.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
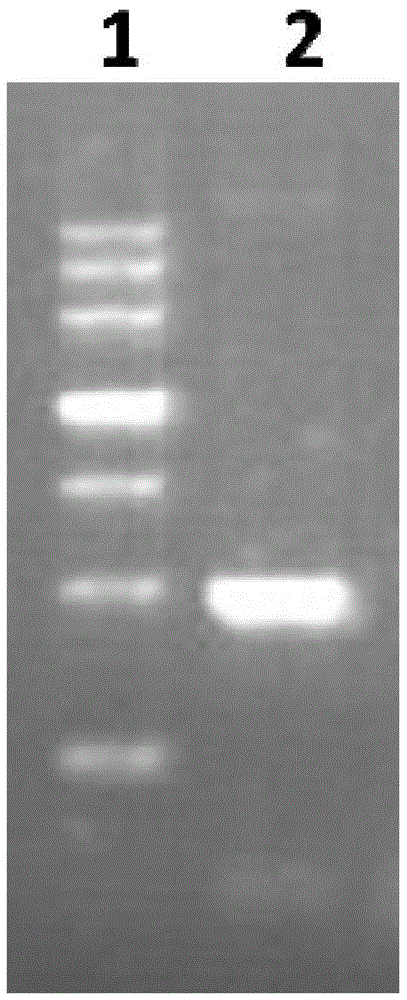
[0064] Embodiment 1: Cloning of OprL gene and construction of recombinant plasmid pGEX-6P-2-OprL
[0065] 1. Firstly, according to the full-length gene sequence of the OprL protein of Pseudomonas aeruginosa PA01, bioinformatics software was used for structural analysis to determine the OprL target gene fragment to be amplified.
[0066] 2. According to the analysis results, the PCR method was used to amplify the OprL target gene fragment from the PA01 genome, and the amplification steps were as follows:
[0067] 1) Design the PCR primers as follows, which are SEQ ID NO: 3-4 (the base sequence of the restriction site is underlined)
[0068] Seq ID
Primer name
Sequence (5'-3')
Seq ID 3
OprL-F
CGC GGATCC TGCTCCTCCAAGGGCG
Bam H I
Seq ID 4
OprL-R
CCG GAATTC TTACTTCTTCAGCTCGACG
EcoR I
[0069] 2) Take out the preserved Pseudomonas aeruginosa strain PA01 from the -80°C freezer and spread it on LB s...
Embodiment 2
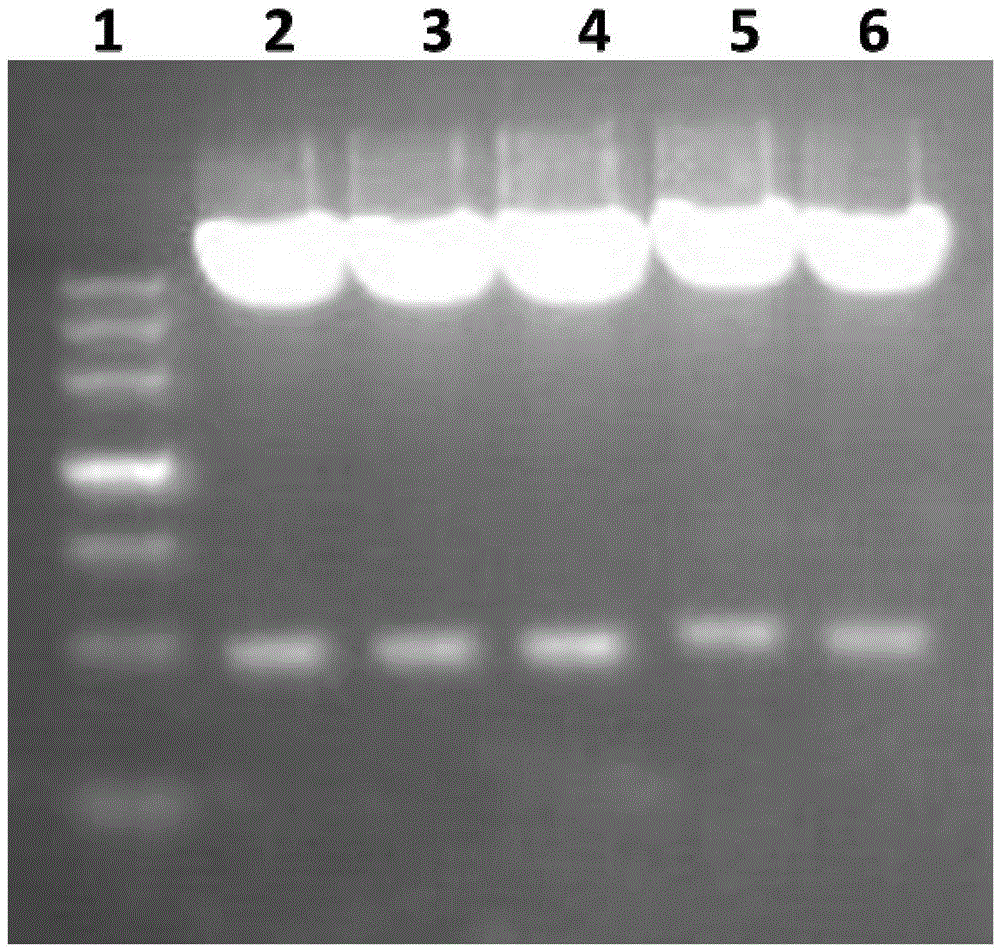
[0098] Example 2: Identification of recombinant fusion protein Vac9 induced expression and expression form in prokaryotic expression system-Escherichia coli
[0099] 1. Induced expression of recombinant protein Vac9
[0100]Take 100 μL of the overnight cultured pGEX-6P-2-OprL / XL-1blue bacterial solution and add it to 10 mL of Amp+ resistant LB medium, culture overnight at 180 rpm at 37°C, respectively take 400 μL of the overnight cultured bacterial solution and add it to 20 mL of Amp+ resistant LB medium (The rest of the bacterial solution is stored in a refrigerator at 4°C for later use), culture at 37°C for 2-3 hours, rotate at 200 rpm, and reactivate until the OD600 is 0.8-1.0, add 4 μL of IPTG to make the final concentration 200 μM, and place on a shaker Induced expression Induced expression at 30°C for 3h.
[0101] 2) Take out the bacterial solution after induced expression, centrifuge at 12000rpm for 5min, discard the supernatant, add 1mL of lysisbuffer (20mMPB, pH7.2, ...
Embodiment 3
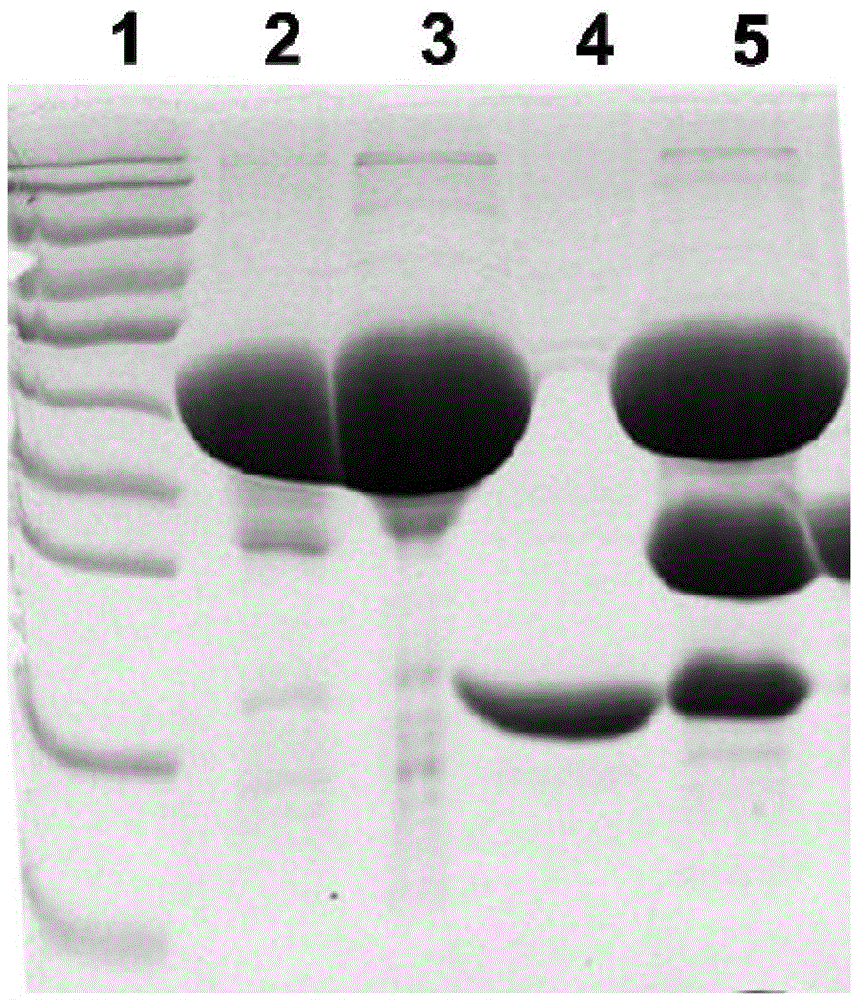
[0108] Embodiment 3: Preparation of recombinant protein Vac9 antigen
[0109] 1. Amplify culture to obtain protein
[0110] Take 400 μL of the pGEX-6P-2-OprL / XL-1blue bacterial solution stored in the refrigerator at 4°C and add it to 20mL LB medium containing Amp resistance for one activation. After culturing at 200rpm for 5-6 hours at 37°C, take 8mL once The activated bacterial solution was added to 400mL LB medium containing Amp resistance for secondary activation, cultured at 37°C for 3-4h until the OD600 was 1.0, then added 80μLPTG (final concentration 200μM) and placed in a shaker at 16°C overnight to induce Afterwards, 12000rpm centrifugal 15min collects thalline, adds 20mLlysisbuffer (same as embodiment 2) after resuspending thalline, bacterium liquid is carried out ultrasonic cracking 3min (200V), collect supernatant and GlutathioneSepharose4B (GE Company) gel beads (beads) binding treatment; then SDS-PAGE gel electrophoresis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com