Nucleoside phosphorylase, encoding gene and high-producing strain and application of nucleoside phosphorylase
A nucleoside phosphorylase high-yield, nucleoside phosphorylase technology, applied in application, genetic engineering, plant genetic improvement and other directions, can solve the problems of low conversion rate and high cost of nucleoside drugs, and achieve easy purification and good application. Potential, high substrate conversion effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] This example illustrates the biological properties and identification of Brevibacillus borstelensis LK01 producing nucleoside phosphorylase.
[0026] A nucleoside phosphorylase-producing strain LK01 was obtained from the organic solvent-resistant bacterial library in our laboratory.
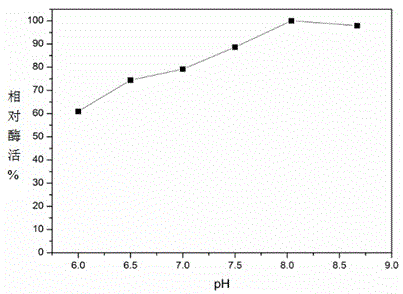
[0027] Biological properties of the strain LK01: the strain is a Gram-positive bacterium, the morphological characteristics of the bacteria are rod-shaped, the cell size is (0.3-0.4) μm*(2-7) μm, there are obvious depressions, and there are flagella around. Mesozoic spores, movement. The colonies are round, milky white in color, smooth, opaque, and not concave. Oxidase reaction was negative and gelatin reaction was positive. The starch hydrolysis test was negative, the nitrate reaction was positive, the VP reaction was negative, the glucose reaction was positive, the citric acid reaction was positive, the arabinose reaction was negative, and the xylose reaction was negative.
[0028] Af...
Embodiment 2
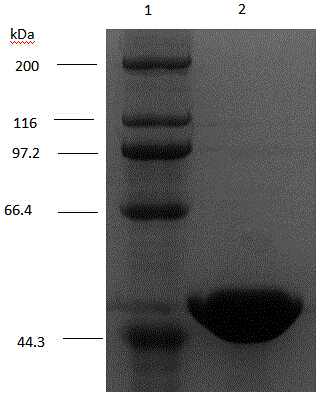
[0030] The nucleoside phosphorylase produced by Brevibacillus borstelensis LK01 is named as nucleoside phosphorylase PyNP.
[0031] This example illustrates the isolation and cloning procedure of the gene encoding nucleoside phosphorylase PyNP.
[0032] Total DNA was extracted by the phenol-chloroform method. According to the analysis of the whole gene sequencing results of Brevibacillus borstelensis (NCBI Reference Sequence: I532_RS05705), a gene encoding pyrimidine nucleoside phosphorylase was obtained, and primers SF and SR were designed according to the gene sequence.
[0033] The sequence of SF (SEQ ID NO: 3) is: ACGGAGCTCGAATTC G GATCC ATGCGCATGGTCGATATCATTG.
[0034] The sequence of SR (SEQ ID NO: 4) is: CGCGGCAGCCATATG G CTAGC CTATTCGGTTATGATTTTGTAAATTAATGG.
[0035] Among them, the underlined part of the primer SF is the NheI restriction site, and the underlined part of the primer SR is the BamhI restriction site.
[0036] Using the genome of Brevibacillus bo...
Embodiment 3
[0038] This example illustrates the preparation of recombinant expression vectors and recombinant bacteria.
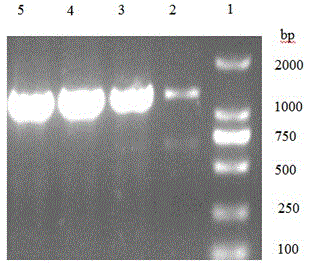
[0039] The nucleoside phosphorylase PyNP gene fragment obtained in Example 2 was double-digested with restriction endonucleases BamHI and NheI at 37°C for 12 hours, purified by agarose gel electrophoresis, and the target fragment was recovered using an agarose gel DNA recovery kit . Under the action of T4 DNA ligase, the target fragment was ligated with the plasmid pET28a digested by BamHI and NheI overnight at 16°C to obtain the recombinant expression plasmid pET-PyNP.
[0040] Transform the recombinant expression plasmid pET-PyNP into Escherichia coli E.coliBL21(DE3) competent cells, screen the positive recombinants on the resistance plate containing kanamycin, select single clones, and verify the colonies by PCR Positive clone means obtaining positive recombinant bacteria E. coli BL21(DE3) / pET-PyNP.
[0041] Inoculate the recombinant strain E.coliBL21(DE3) / pET-PyN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com