Wild porcine pseudorabies strain and gene-deleted strain dual real-time fluorescence quantification PCR detection kit, primers and probe
A technology of real-time fluorescence quantification and porcine pseudorabies, which is applied in the direction of microbial determination/testing, microbial-based methods, biochemical equipment and methods, etc., to achieve the effect of simple operation, ensuring safety and rationality, and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
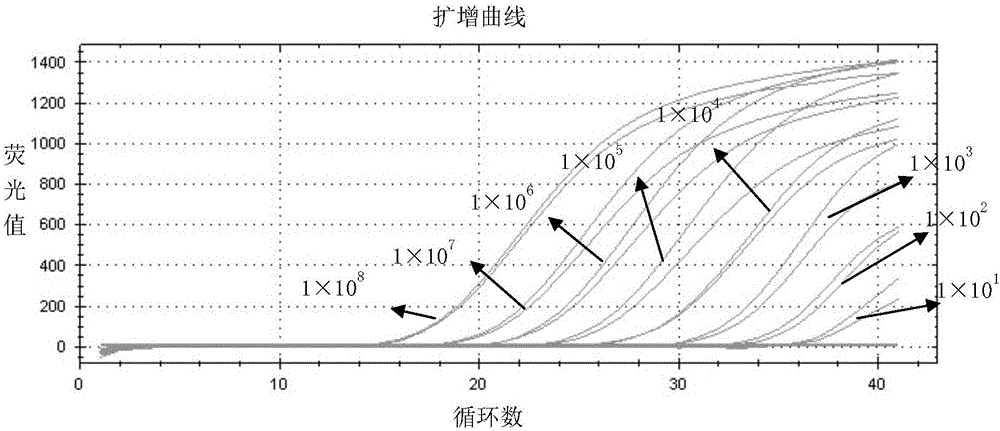
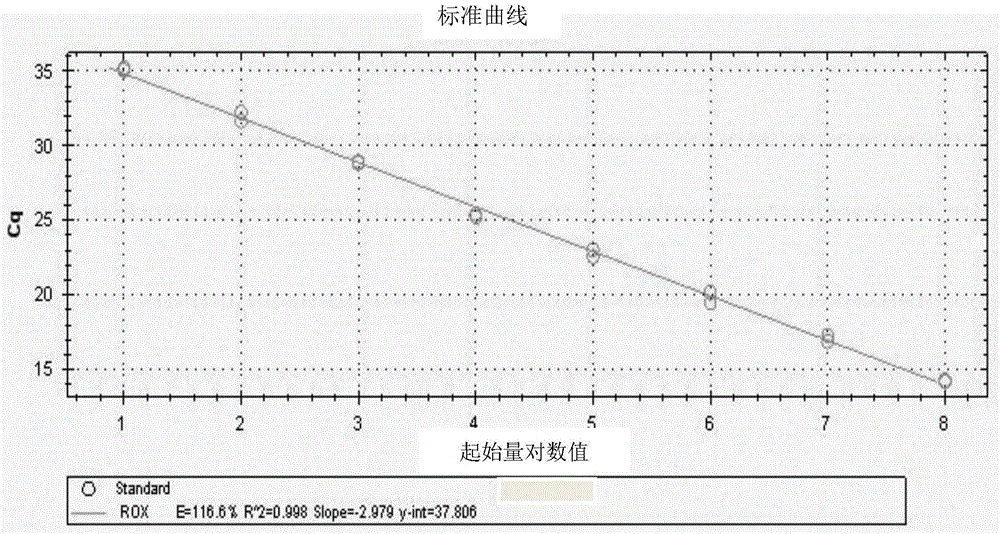
[0044] Example 1. Design of primers and TaqMan probes for dual real-time fluorescent quantitative PCR detection of gB gene and gE gene of porcine pseudorabies wild strain and gene deletion strain
[0045] The sequence of gB gene (GenBank number: NC_006151) and gE gene (GenBank number: AF207700) of porcine pseudorabies wild strain was retrieved from NCBI's nucleic acid database GenBank (http: / / www.ncbi.nlm.nih.gov), After comparison with DNA Man software, according to the principles of primer design, 3 pairs of primers were designed for the gB gene and the gE gene respectively for PCR amplification, and the primers with high primer specificity and good amplification effect were selected by electrophoresis results. Yes, the primer pair sequence to determine the specificity of the gB gene is the 17211-17574th base from the 5' end, and the primer pair sequence specific to the gE gene is the 443-1357th base from the 5' end; Using Primer Express 6.0 software, primers and TaqMan prob...
Embodiment 2
[0058] Embodiment 2, using the primers of the present invention and TaqMan probes to carry out double real-time fluorescent quantitative PCR detection on the gB gene and gE gene of the porcine pseudorabies wild strain and the gene deletion strain
[0059] 1. Extract the genomic DNA of the sample to be tested
[0060] Genomic DNA was extracted from the cell cultures of inactivated porcine pseudorabies field strains and gene-deleted strains and 12 collected clinical suspected serum disease materials. The specific method included the following steps:
[0061] (1) Take 200 μL of inactivated pseudorabies field strain and gene deletion strain cell culture and clinical serum into 1.5mL centrifuge tube, add 1mL DNAzol lysate, shake and mix, let stand for 10 minutes to lyse;
[0062](2) Centrifuge at 12000 rpm for 10 minutes, take about 800 μL of supernatant into a new 1.5 mL centrifuge tube, add 500 μL of absolute ethanol, mix gently and let stand for 5 minutes to precipitate DNA;
...
Embodiment 3
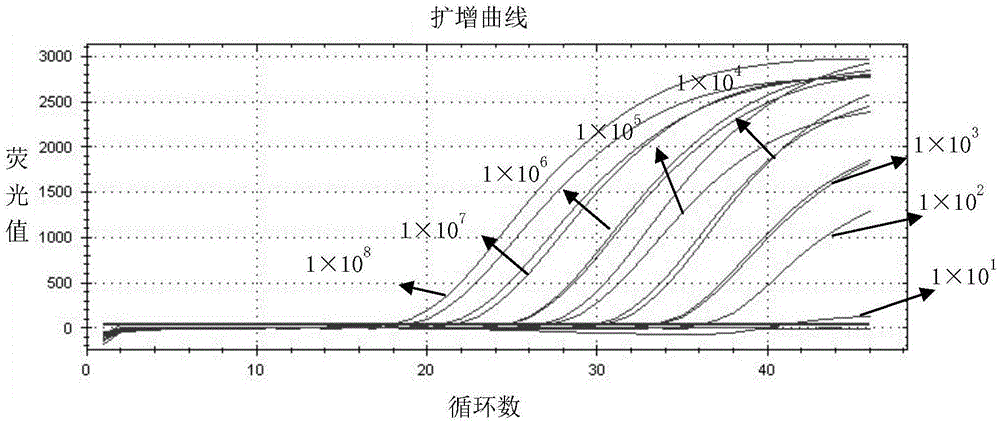
[0087] Embodiment 3, the specificity experiment of detection method of the present invention
[0088] According to the method described in embodiment 2, extract RNA to swine fever virus (CSFV), bovine viral diarrhea virus (BVDV) and carry out reverse transcription, directly to porcine circovirus type 2 (PCV2), rhinotracheitis virus (IBR) DNA was extracted, while the inactivated porcine pseudorabies wild strain cell culture was used as a positive control, and enzyme-free water was used as a negative control. Under the guidance of primers and TaqMan probes of the present invention, double real-time fluorescent quantitative PCR detection was carried out. PCR reaction The system and reaction conditions refer to Example 2 to verify the specificity of the method.
[0089] Test results such as Figure 5 As shown, the CT values of the positive control gB gene and the gE gene were 20 / 21 (38, and no specific amplification curve appeared, and the results were negative. The detection ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com