Fluorescent quantitative PCR (polymerase chain reaction) primer for rapidly diagnosing nosema antheraeae and application of primer
A fluorescence quantitative and microsporidia technology, which is applied in the direction of microorganism-based methods, microorganism measurement/inspection, microorganisms, etc., can solve the problems of low detection sensitivity, cumbersome operation, and late detection period of the tussah silkworm microsporidia diagnostic technology. Achieve the effect of eliminating false positives in molecular diagnosis, quantifying reliability, and improving yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
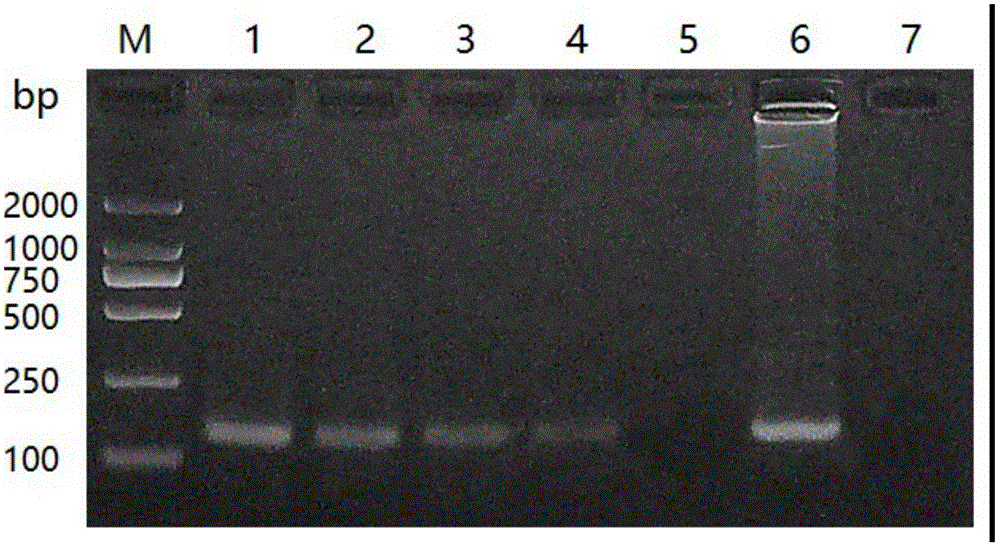
[0041] The specificity of embodiment 1 conventional PCR identification primer
[0042] Take the tussah microsporidia suspension, centrifuge at 5000r / min for 5min, remove the supernatant and retain the spore precipitate, add the extract (10mmol / L Tris-HCL pH 8.0; 10mmol / L EDTA pH 8.0; 100mmol / L NaCl; 2% SDS; 0.039 mol / L DTT), plus proteinase K (final concentration: 100 μg / mL), digested at 55°C for 2 hours, extracted with phenol / chloroform, and obtained the genomic DNA of Nos. tussah mori as a positive reference.
[0043] Take another tussah silkworm chrysalis, tussah silkworm larvae and tussah silkworm mother moth infected with microparticles disease, take appropriate amount of tussah silkworm pupae tissue, tussah silkworm larvae tissue, tussah silkworm moth tissue and tussah silkworm eggs (taken from the body of tussah silkworm moth), and put them into No. 1# respectively. In the 1.5mL EP tubes of 2#, 3# and 4#, take an appropriate amount of healthy tussah silkworm chrysalis t...
Embodiment 2
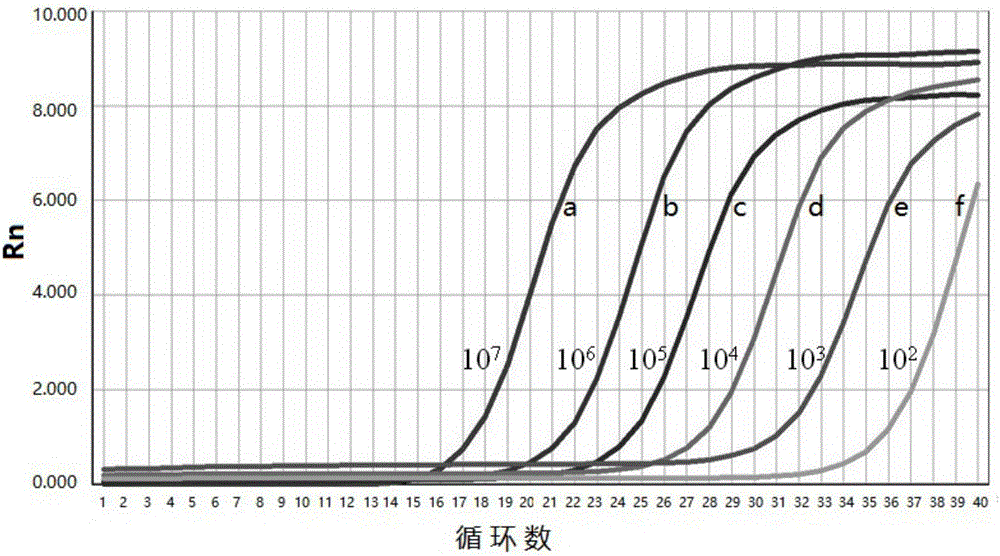
[0049] Standard curve and primer amplification efficiency of embodiment 2 fluorescence quantitative PCR
[0050] Preparation of standards:
[0051] The positive reference PCR product in Example 1 was recovered and purified, connected to the pMD-18T vector, transformed into DH5α competent cells, spread on a plate medium for cultivation, and picked positive colonies to extract plasmid DNA for sequencing identification. Quantify the pMD-18T / Np-SSUrRNA plasmid DNA with correct sequencing results, convert it into copy number, and dilute it in a 10-fold gradient with sterile water to 1×10 8 ~1×10 1 copies / μL of the standard.
[0052] Drawing of standard curve and determination of detection limit and primer efficiency:
[0053] ABI 7300 fluorescent quantitative PCR instrument was used for amplification. Fluorescence quantitative PCR reaction system: SYBR PremixExTaq 10 μ L, ROX Reference Dye I 0.4 μ L, the upstream primer (10pmol / μL) 0.2 μ L as described in SEQ ID NO:1, the downs...
Embodiment 3
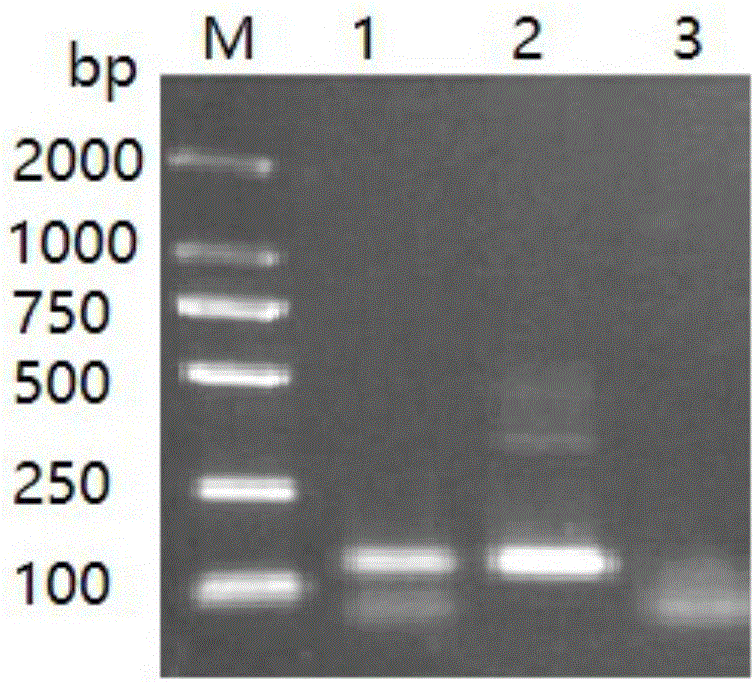
[0055] Embodiment 3 Fluorescent quantitative PCR detects Microsporidia tussah
[0056] Take tussah silkworm chrysalis, tussah silkworm larvae and tussah silkworm mother moth infected with microparticles disease, take appropriate amount of tussah silkworm pupae tissue, tussah silkworm larvae tissue, tussah silkworm moth tissue and tussah silkworm eggs (taken from the body of tussah silkworm moth), and put them into No. 1 and 2 respectively. In #, 3# and 4# 1.5mLEP tubes, take an appropriate amount of healthy tussah silkworm chrysalis tissue into the negative reference EP tube, add nucleic acid extraction solution (10mmoL / L Tris-HClpH8.0, 100mmoL / L EDTA, 0.5% SDS, 100μg / mL proteinase K), digested at 55°C for 4 hours, extracted with phenol / chloroform, and obtained the DNA of each sample.
[0057] For 1-4# sample DNA, deionized water, healthy pupae DNA and positive reference as template DNA, ABI 7300 fluorescent quantitative PCR instrument was used for fluorescent quantitative PC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com