Application of OsSAPK7 protein and coding genes thereof in improving resistance to bacterial blight of rice
A technology for transgenic rice and leaf blight, which can be used in application, genetic engineering, plant genetic improvement and other directions, and can solve problems such as difficulty in using disease resistance genes and loss of variety resistance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
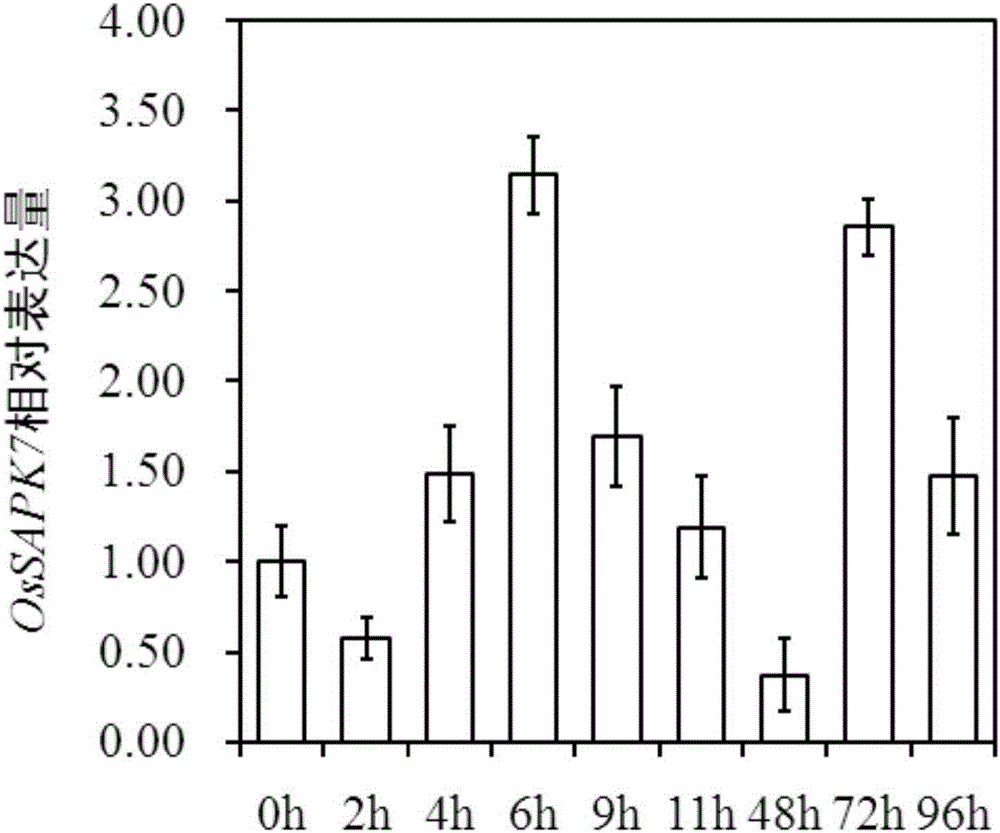
[0065] Embodiment 1, OsSAPK7 gene expression analysis
[0066] 1. Sow the seeds of 9804 rice in seedling trays filled with sterilized soil, cultivate them in the greenhouse for 25 days, and then transplant them into the net room for planting as a single plant.
[0067] 2. At the tillering stage of the rice plants in step 1, use the Xoo strain ZHE173 to artificially inoculate the rice plants by the leaf-cutting method, and inoculate 5 leaves per plant.
[0068] 3. Complete the 0, 2, 4, 6, 9, 11, 48, 72, and 96 hours of manual inoculation in step 2, cut the inoculated leaves, quickly put them into liquid nitrogen for quick freezing, and take three biological replicates for each sample. All samples were frozen in liquid nitrogen and stored at -70°C.
[0069] 4. Take the sample obtained in step 3, extract the total RNA of the sample, and reverse transcribe it into cDNA.
[0070] 5. Using the cDNA obtained in step 4 as a template, carry out qRT-PCR reaction, use primer F and prim...
Embodiment 2
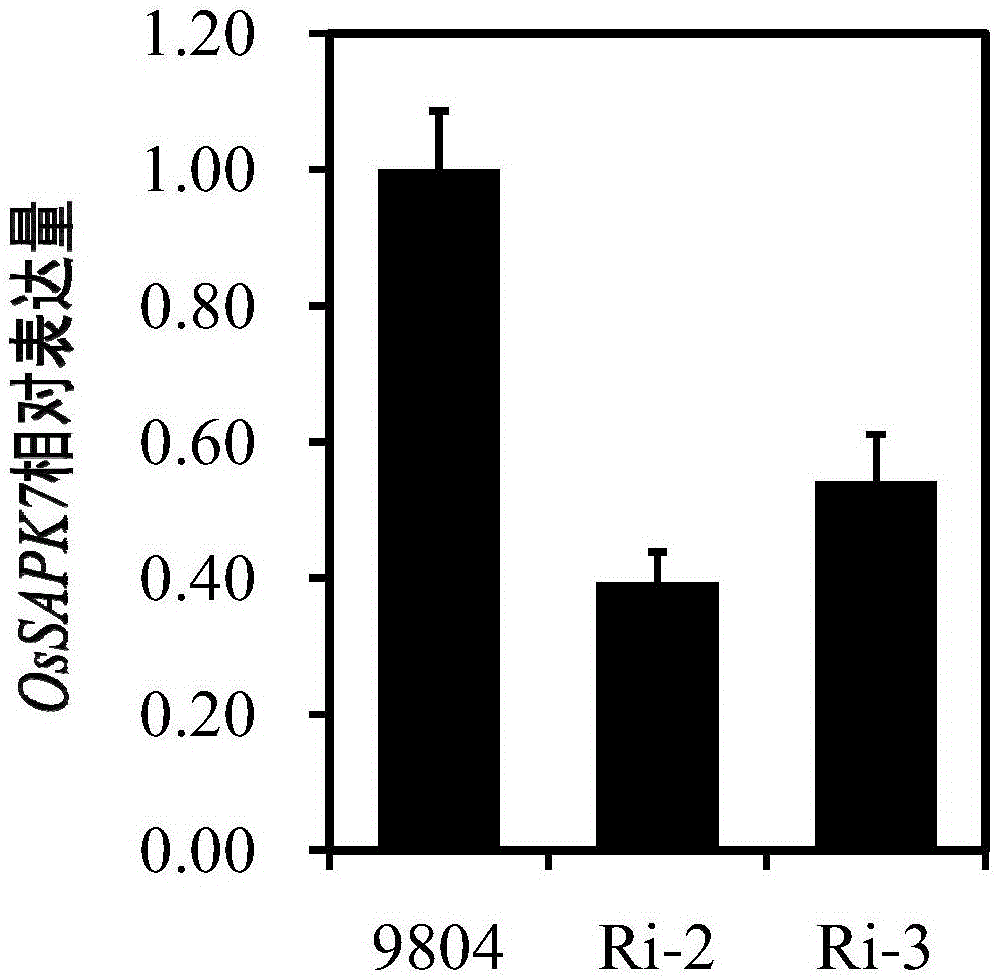
[0078] Embodiment 2, OsSAPK7 gene functional analysis
[0079] 1. OsSAPK7 Gene RNAi Vector Construction
[0080] 1. Extract the total RNA from 9804 rice leaves and reverse transcribe it into cDNA.
[0081] 2. Using the cDNA obtained in step 1 as a template, PCR amplification was performed with primers attB-F and primers attB-R to obtain amplified products.
[0082] attB-F: 5′- GGGGACAAGTTTGTACAAAAAAGCAGGCT GGCAGCTGATGGTCACACT-3′:
[0083] attB-R: 5′- GGGGACCACTTTGTACAAGAAAGCTGGGT AGATGCTCAATGAGATGGTTTT-3'.
[0084] 3. Through the BP reaction, the amplified product obtained in step 2 was introduced into the vector pDONR201 to obtain a positive entry clone plasmid pDONR201-OsSAPK7i (which has been verified by sequencing) containing the double-stranded DNA molecule shown in sequence 3 of the sequence listing.
[0085] BP reaction system: amplification product 2.7μL (50-100ng), vector pDONR2011.0μL (30-50ng), 5×BPReaction Buffer 1.0μL, BP Enzyme mix 0.3μL.
[0086] BP reacti...
Embodiment 3
[0115] Embodiment 3, the application of OsSAPK7 gene in improving rice bacterial blight
[0116] 1. Construction of OsSAPK7 gene overexpression vector
[0117] 1. Extract the total RNA from 9804 rice leaves and reverse transcribe it into cDNA.
[0118] 2. Using the cDNA obtained in step 1 as a template, PCR amplification was carried out with primers attB-F1 and primer attB-R1 to obtain amplified products.
[0119] attB-F1:5′- GGGGACAAGTTTGTACAAAAAAGCAGGCTGC ATGGAGAGGTACGAGCTGCTC-3′:
[0120] attB-R1:5′- GGGGACCACTTTGTACAAGAAAGCTGGGT TCAGCTGAGCTGAAACTCACCA-3'.
[0121] 3. Through the BP reaction, the amplified product obtained in step 2 was introduced into the vector pDONR201 to obtain the positive entry clone plasmid pDONR201-OsSAPK7 containing the double-stranded DNA molecule shown in Sequence 1 of the Sequence Listing.
[0122] BP reaction system: amplification product 2.7μL (50-100ng), vector pDONR201 1.0μL (30-50ng), 5×BPReaction Buffer 1.0μL, BP Enzyme mix 0.3μL. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com