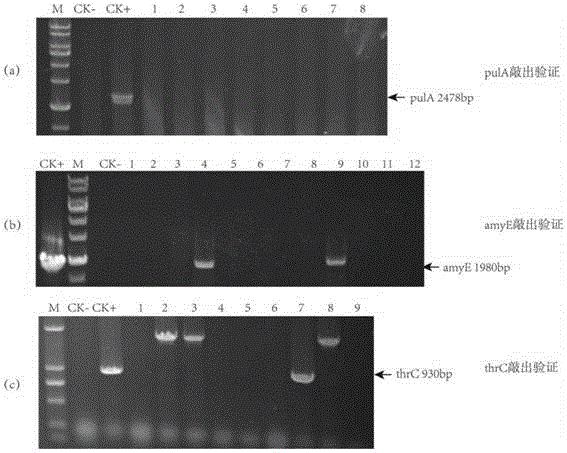
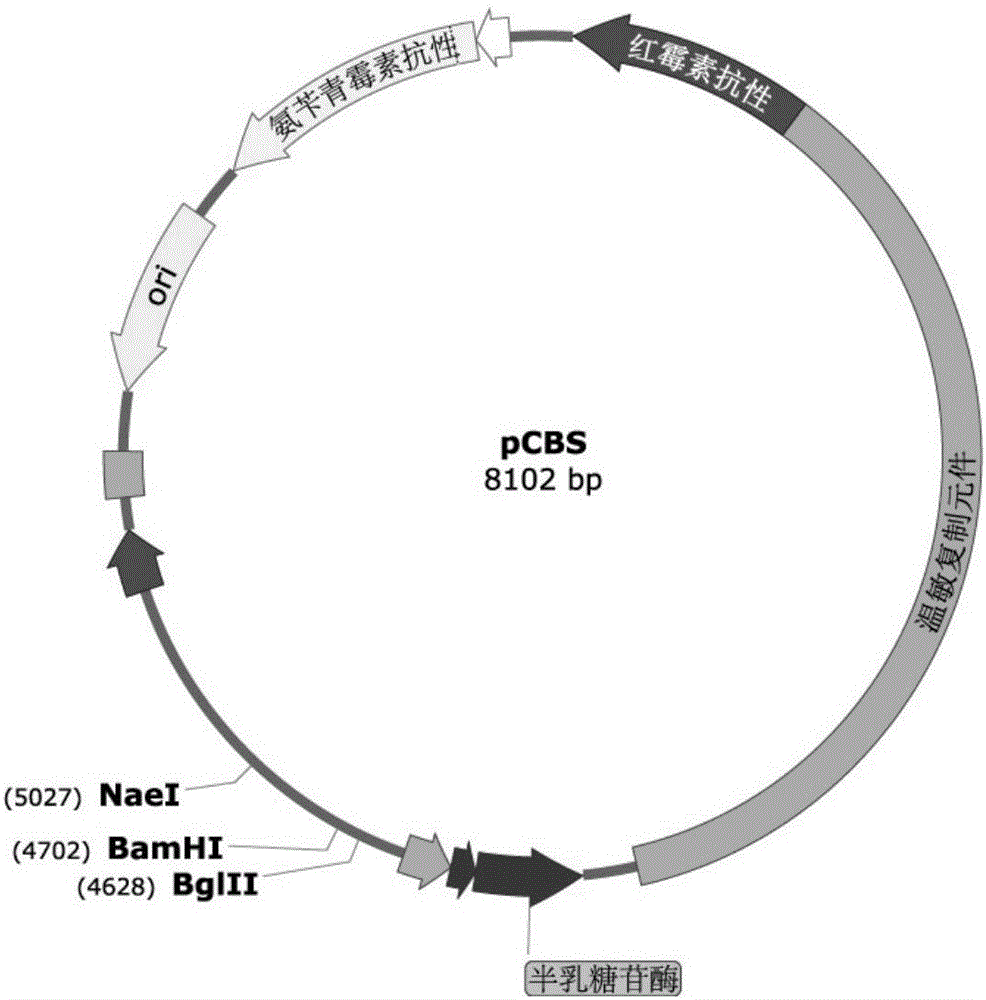
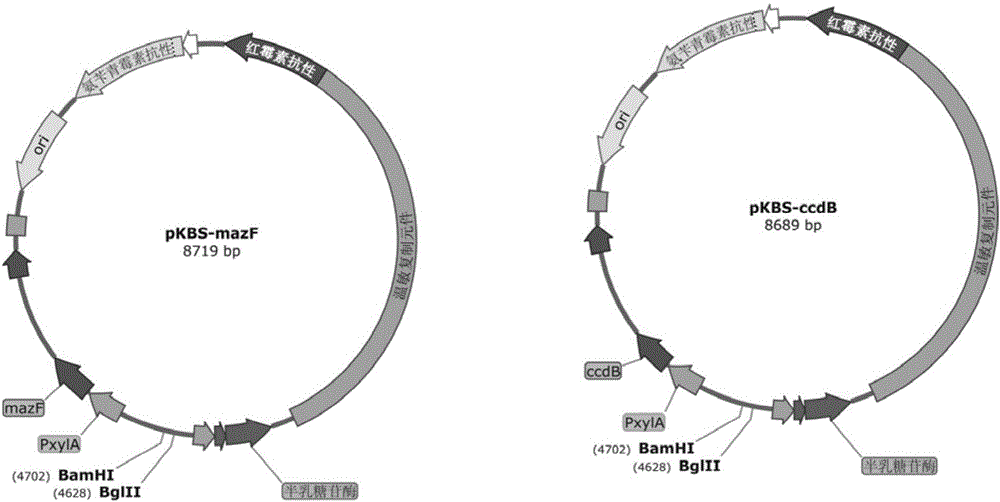
Marker protein expression cassette capable of inducing regulation and recombinant vector constructed by same and application
A technology for labeling proteins and recombinant vectors, applied in the field of molecular biology, can solve the problems of low transformation efficiency of Bacillus, inability to overcome recombination efficiency, and inability to obtain, etc., to achieve the effect of increasing the quantity, reducing the difficulty of operation, and improving the success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] In this embodiment, the grac promoter (lactose operon contained) and the toxin protein ccdB recombinant plasmid containing optimized codons were selected. Using the genome of BS001 as a template, through
[0046] Primer 49: acacattaactagacagatctTTTGCCGAAGCTGATCCG (SEQ ID NO.57)
[0047] and primer 50: atggcatgcatcgatagatctGAAGTGTTACCTGTTGACGCGC (SEQ ID NO. 58)
[0048] Amplification of the homology arm upstream of thrC (SEQ ID NO.69)
[0049] Pass Primer 51: acacattaactagacagatctCGAAGCCGACATGCTGTTCT (SEQ ID NO. 59)
[0050] and primer 52: atggcatgcatcgatagatctTTTTCTAAATAAAAACTCCTTTATCTCCA (SEQ ID NO.60) to amplify the homology arm (SEQ ID NO.70) downstream of thrC (PCR conditions: high-fidelity pfu enzyme 1 μL, 10×PCRbuffer 2.5 μL, dNTP (10 mmol / L) 2 μL, Primer (10 μmol / l) 2 μL each, template (Bacillus subtilis BS001 DNA) 1 μL, double distilled water 14.5 μL. Gently blow and mix with a pipette tip. Pre-denaturation at 95°C for 3 minutes; denaturation at 95°C for 40 s...
Embodiment 2
[0052]In this embodiment, the grac promoter (lactose operon contained) and the toxin protein ccdB recombinant plasmid containing optimized codons were selected. Using the genome of BS001 as a template, the homology arm upstream of thrC was amplified by primer 49 and primer 50, and the homology arm downstream of thrC was amplified by primer 51 and primer 52 (the amplification process was the same as in Example 1). pKBS was digested with restriction endonuclease BglⅡ and connected with the upstream homology arm of thrC. Ligated with the downstream homology arm of thrC by restriction endonuclease BamHI digestion. Using pet30A as a template, through
[0053] Primer 53: tgcaaaagccgcagcagatctATGAGCCATATTCAACGGGAAA (SEQ ID NO. 61) and
[0054] Primer 54: atggcatgcatcgatagatctTTAGAAAAACTCATCGAGCATCAAA (SEQ ID NO.62) amplifies the kanamycin resistance gene kan (PCR conditions: high-fidelity pfu enzyme 1 μL, 10×PCR buffer 2.5 μL, dNTP (10 mmol / L) 2 μL, primer (10 μmol / L l) Each 2 μL,...
Embodiment 3
[0056] In this embodiment, the xylose-induced xylA promoter and the toxin protein mazF recombinant plasmid containing optimized codons were selected. Using the genome of BS001 as a template, the homology arm upstream of thrC was amplified by primer 49 and primer 50, and the homology arm downstream of thrC was amplified by primer 51 and primer 52 (the amplification process was the same as in Example 1). Pkbs was digested with restriction endonuclease BglⅡ and connected with the upstream homology arm of thrC. Ligated with the downstream homology arm of thrC by restriction endonuclease BamHI digestion. The vector was then transformed into Bacillus subtilis BS001, and a single colony was obtained on an erythromycin (5 μg / ml) resistant plate, picked and inoculated with LB medium containing erythromycin (5 μg / ml), and treated at 42°C for 12 hours, Erythromycin (5 μg / ml) resistant plates containing X-gal were coated. Pick a blue single colony, inoculate LB medium and culture at 37°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com