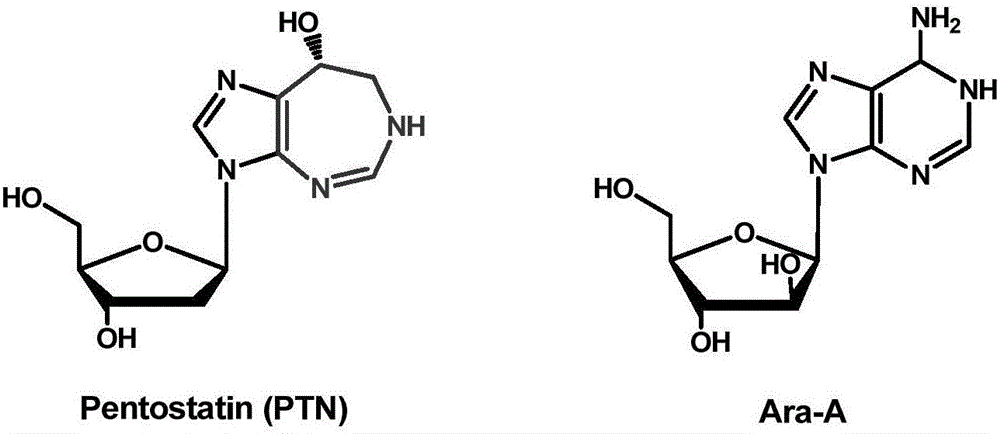
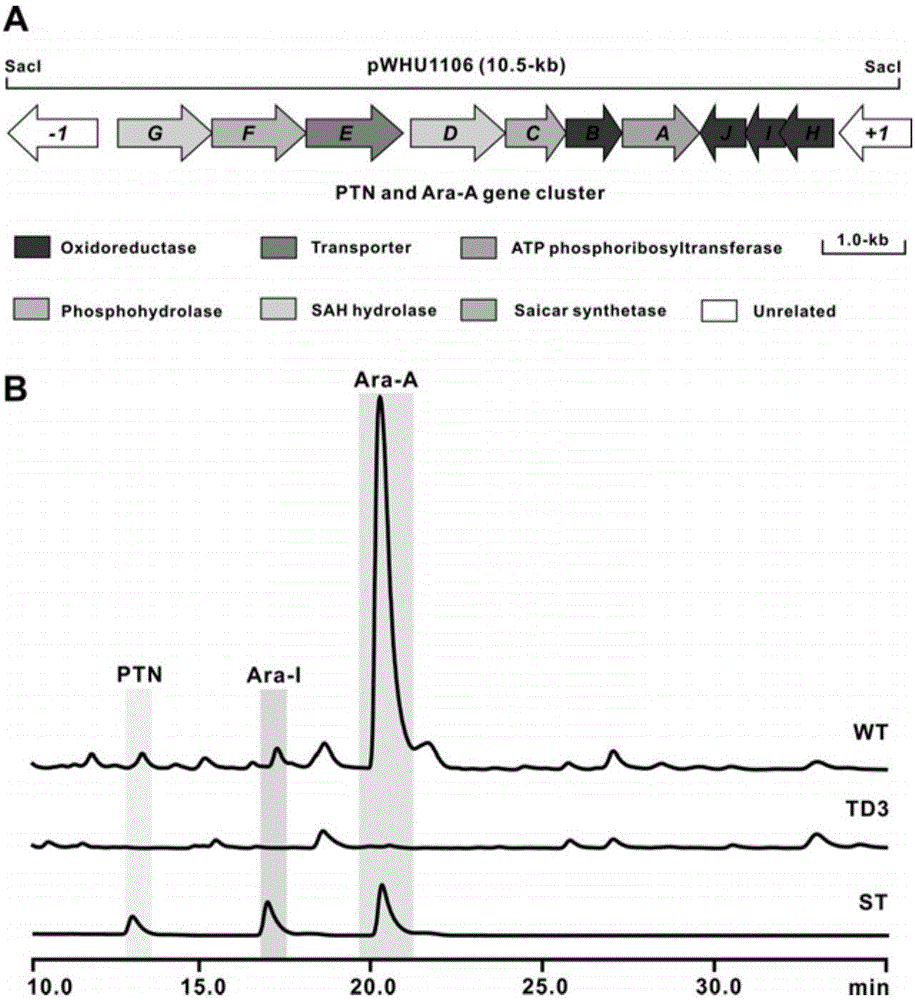
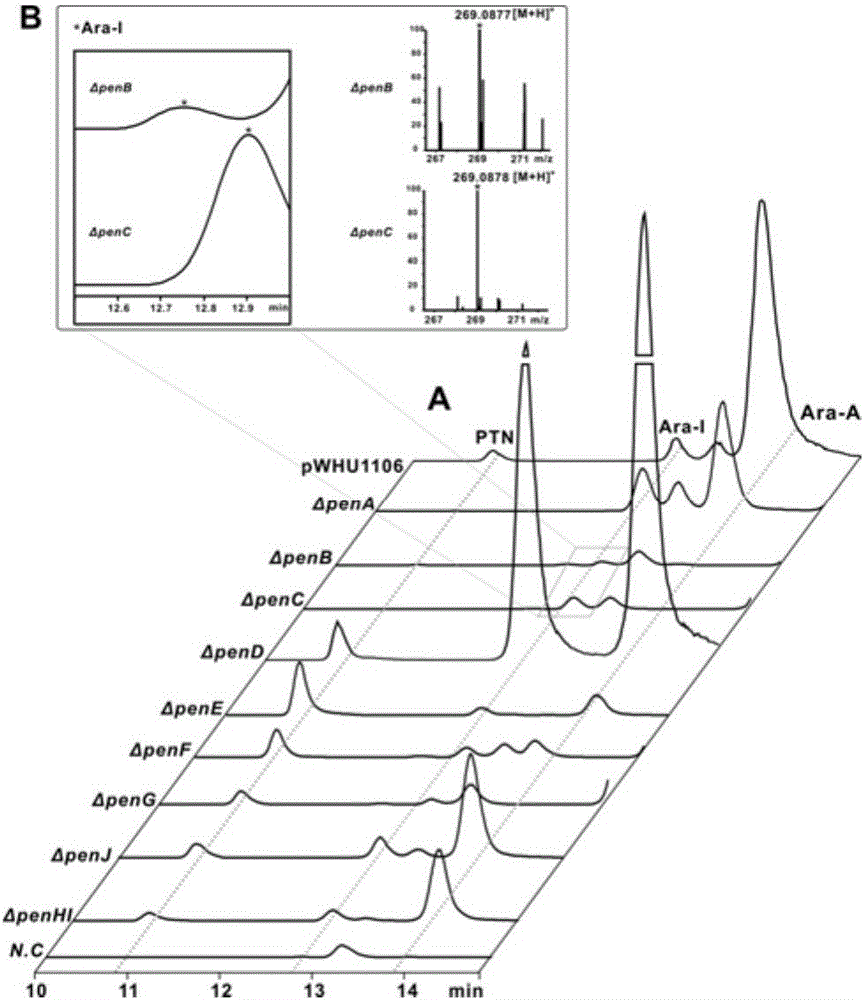
Biosynthetic gene cluster of pentostatin and arabinofuranosyladenine and application of biosynthetic gene cluster
A kind of arabinoside biological and pentostatin technology, applied in the field of microbial gene resources and genetic engineering, can solve the problem of little understanding of biosynthesis and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] [Example 1] Extraction of total DNA of pentostatin and vidarabine-producing bacteria Streptomyces NRRL 3238 total DNA:
[0060] Take 30 μL of Streptomyces NRRL 3238 spores into 50 mL of TSBY medium, culture at 30°C and 200 rpm for 24-36 hours until the medium becomes turbid. 50 mL of Streptomyces NRRL 3238 bacteria solution was centrifuged at 4000 rpm at 4°C for 10 min to remove the supernatant and collect the bacteria. Dissolve the bacteria in 25mL of 10.3% sucrose solution, oscillate and wash the bacteria, centrifuge at 4000rmp, 4°C, 10min to remove the supernatant; then dissolve the bacteria in 15mL set buffer, shake and mix, 4000rmp, 4°C, Centrifuge for 10 minutes to remove the supernatant, and repeat twice; dissolve the bacteria in 10 mL of set buffer, shake and mix well, add 50 μL of lysozyme solution (100 mg / mL) and place it in a 37°C water bath for 30 minutes; then add 280 μL of protease K solution (50mg / mL), after mixing, add 600μL of 10% SDS, mix it upside do...
Embodiment 2
[0061][Example 2] Establishment of Genomic Library of Streptomyces NRRL 3238 Genomic Library of Pentostatin and Vidarabine Producing Bacteria
[0062] First, determine the amount of Sau 3AI through a series of dilution experiments, prepare a total volume of 500 μL enzyme digestion system (5 μL 0.1u / μL Sau 3AI, 495 μL 10Xbuffer diluted DNA) solution 1, take 300 μL solution 1 and mix 100 μL DNA To form solution 2 (Sau 3AI final concentration 0.075u / 100μL), take 200μL solution 1 and 200μL DNA and mix to form solution 3 (Sau 3AI final concentration 0.05u / 100μL), take 200μL solution 2 and 200μL DNA and mix to form solution 4 ( Sau 3AI final concentration 0.0375u / 100 μL), take 200 μL solution 4 and 200 μL DNA and mix to form solution 5 (Sau 3AI final concentration 0.01875u / 100 μL). Mix the above solutions evenly and put them on ice, then put them in a water bath at 37°C for 1 hour, take them out and put them on ice immediately, and electrophoresis on 1% agarose gel with a length of ...
Embodiment 3
[0068] [Example 3] Fermentation conditions of pentostatin and vidarabine producing bacteria Streptomyces NRRL 3238, product high performance liquid phase (HPLC) detection conditions
[0069] The spores of Streptomyces NRRL 3238 were inoculated into the seed medium, cultured at 28°C, 220rmp for 48h, transferred to a fermentation shaker flask at 4% inoculum size, and cultured at 28°C, 220rmp for 6 days. Collect the fermentation broth, centrifuge the fermentation broth at 12000rmp for 20min, take the supernatant and pass it through a 0.22μm microporous membrane for detection and analysis by HPLC and LC-MS.
[0070] HPLC detection conditions: phase A is ultrapure water added with 0.15% trifluoroacetic acid (TFA), and phase B is methanol. The initial 95% phase A gradient eluted to 80% within 30min, and phase A was converted to 10% at 31min and maintained at this concentration for 45min, and phase A was converted to 95% at 46min and maintained for 65min. The flow rate is 0.5mL / min,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com