A molecule adaptor and applications thereof
A molecular linker and sequence technology, applied in the field of sequencing, can solve the problems of high sequencing background noise, complicated experimental process, and low effective data rate, and achieve the effects of increased detection sensitivity, accurate detection results, and simple preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Joint annealing and extension steps
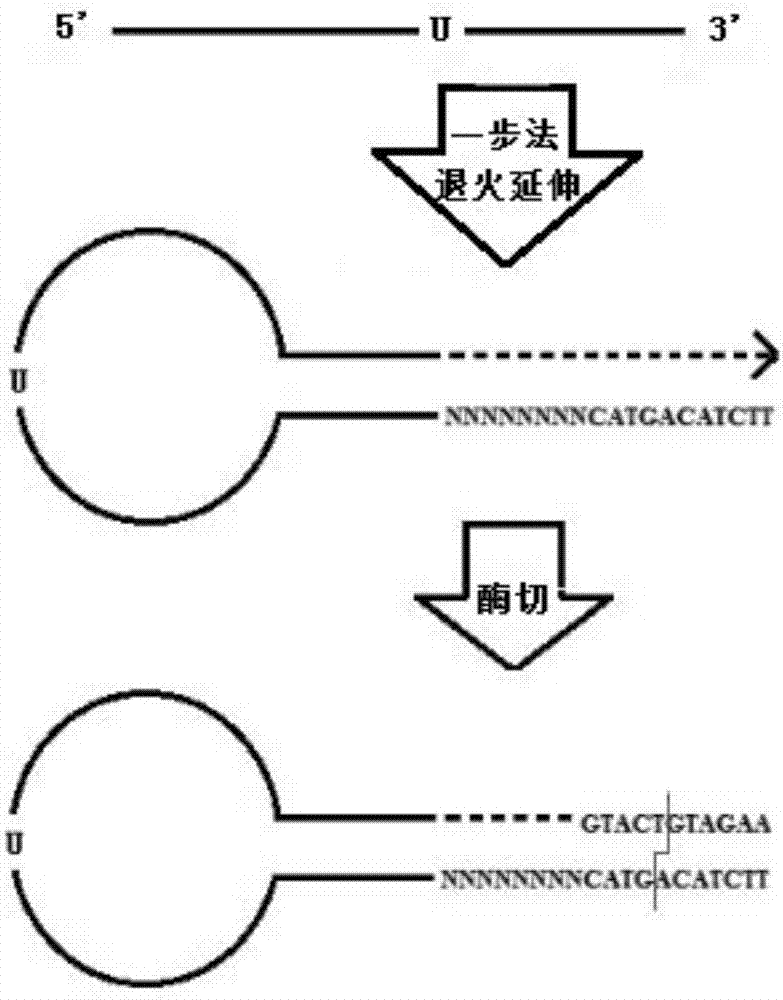
[0049] (1) The sequence of the key-shaped molecular linker is SEQ ID No.1 ( figure 1 ):
[0050] PHO-5’-TTCTACAGTACNNNNNNNN AGATCGGAAGAG CACACGTCTGAACTCCAGTCACdUACACTCTTTCCCTACACGACG CTCTTCCGATC*T -3...
[0051]Note: PHO stands for phosphorylation at the 5' end, where N stands for any base in A / T / G / C, dU stands for deoxyuracil, the left and right underlines of dU are complementary regions, * stands for sulfur modification, dotted line "… ..." represents the extension region, and the italic part is the restriction endonuclease recognition region.
[0052] (2) Reagents required for one-step annealing extension of key-shaped joints:
[0053] Linker sequence (synthesized by Jinweizhi Biotechnology Co., Ltd.), KAPA HiFi Hotstat ReadyMix (KAPA company kk2602), sterilized ultrapure water
[0054] (3) The key-shaped joint adopts one-step annealing and extension steps:
[0055] The synthesized dry powder linker sequence wa...
Embodiment 2
[0069] Example 2 DNA library construction of plasma and tissue samples
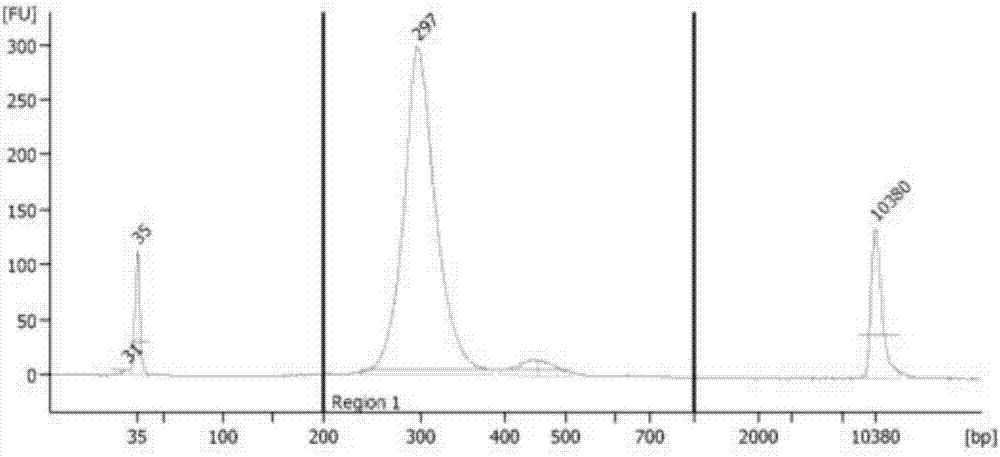
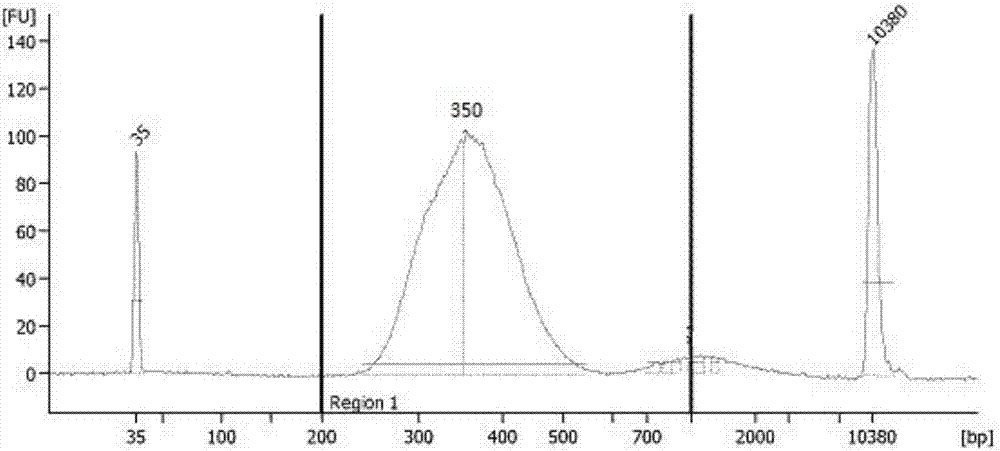
[0070] The samples in this example come from the General Hospital of Shenyang Military Region, 5 patients with clinically diagnosed stage III adenocarcinoma of lung cancer. The plasma (2ml) and tissue samples were taken before the preoperative medication, and the free DNA (cfDNA) and tissue DNA were extracted. The tissue DNA was ultrasonically analyzed. Fragmented into a size of 150-250bp, cfDNA and tissue fragmented DNA were qualified by the Agilent 2100 bioanalyzer, and the libraries were constructed according to the following steps.
[0071] (1) Sample DNA end repair
[0072] Configure the mixed reaction according to Table 4, use the KAPA LTP Library Preparation Kit (KK8233) EndRepair, put all the plasma cfDNA, and input 100ng of the fragmented DNA sample.
[0073] Table 4. Sample DNA end repair system
[0074] Fragmented DNA sample (150bp) 50ul KAPA End Repair Buffer(10X) 7ul K...
Embodiment 3
[0097] Practical Example 3 Cellular DNA Sensitivity Experiment of Known Mutation Sites
[0098] The cell samples used in this example come from the cell bank of the Type Culture Collection Committee of the Chinese Academy of Sciences, including H1975 cell lines (known EGFR L858 and T790M mutations), H1650 cell lines (known EGFR exon 19 deletion), negative MRC cell lines (no EGFR mutation). DNA was extracted from H1975 cells and H1650 cells, and mixed according to the mass ratio of 1:1 after being interrupted by ultrasound, and then blended with negative cell line MRC fragmented DNA samples according to 1%, 0.1%, 0.05%, and 0%, for library construction, and then Two rounds of specific hybridization capture were carried out, and the library after capture was detected by fluorescent quantitative PCR method for corresponding mutation sites, and finally double-end sequencing was performed to judge the detection sensitivity of molecular joints.
[0099] The specific library constru...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com