Method for producing long-chain binary acid by using recombinant escherichia coli strain
A technology for recombining Escherichia coli and long-chain dibasic acids, applied in the direction of microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problems of difficult entry into cells, low yield, etc., to increase yield and overcome reaction Harsh effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Construction of embodiment 1 recombinant strain MJ1
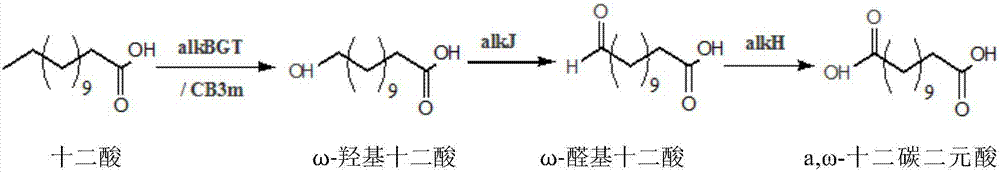
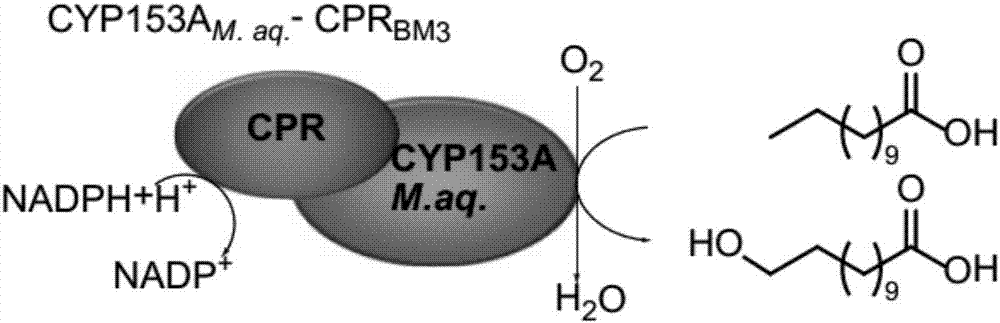
[0027] a, The precursor of ω-dodecanedibasic acid synthesis is dodecanoic acid, but in the cell fatty acid will be in fatty acyl-CoA synthetase encoded by fadD gene, acyl-CoA dehydrogenase encoded by fadE gene and The enoyl-CoA encoded by the fadB gene is gradually degraded through the β-oxidation pathway. Therefore, it is first necessary to block or weaken the β-oxidation pathway genes so that cells can accumulate fatty acids under the expression of other fatty acid synthase genes.
[0028] The wild-type W3110 strain was used as the basic strain to knock out the fadD gene to weaken the fatty acid degradation pathway. The cell construction process was as follows: using pKD4 as a template, the primer pair fadD-K-F / fadD-K-R amplified the knockout fragment (see Table 1 for the primer sequence), The large W3110 electroporation competent cells were electrotransformed, spread on the Kana-resistant plate for screening, and...
Embodiment 2
[0030] Example 2 Construction of long-chain fatty acid synthesis expression plasmid pEZ07-fatB
[0031] According to SEQ ID NO: 1, the acyl-[ACP]thioesterase gene fatB derived from Umbellularia California was synthesized and connected into the pUC57 vector (purchased from Suzhou Jinweizhi Biotechnology Co., Ltd.) to obtain the plasmid pUC57-fatB. The fatB gene is mainly used to catalyze the synthesis of long-chain fatty acids, especially twelve-carbon fatty acids.
[0032] The expression plasmid pEZ07-fatB was constructed by seamless cloning and recombination technology. pEZ07 is a low-copy plasmid with spectinomycin resistance and Trc as a promoter, constructed in our laboratory using pTrc99A and pCL1920 from Biovector China Plasmid Vector Strain Cell Gene Collection Center. Using the plasmid pUC57-fatB synthesized by the above gene as a template, the primer pair fatB-N-F / fatB-X-R amplifies the gene fatB (see Table 1 for the sequence of the primer pair) containing homologous...
Embodiment 3
[0044] Example 3 Construction of long-chain fatty acid synthesis expression plasmid pEZ07-fatB
[0045] The Escherichia coli fadR gene is a regulator of fatty acid synthesis and degradation-related transcriptional genes. The literature shows that overexpressing fadR helps to increase the amount of fatty acid synthesis. Therefore, the fadR gene is co-expressed on the basis of the expression plasmid pEZ07-fatB in Example 1. In order to increase the production of twelve carbon fatty acids.
[0046] The expression plasmid pEZ07-fatB-fadR was constructed by seamless cloning and recombination technology. Using the MG1655 genome as a template, the primer pair fadR-P-F / fadR-H-R amplified the gene fatR with homologous regions at both ends (see Table 1 for the sequence of the primer pair), and recovered it; The recovered plasmid pEZ07-fatB was digested with PstI and HindIII for EZ clone, transformed into Escherichia coli competent cell TG1, and the construction of plasmid pEZ07-fatB-fa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com