Urate oxidase gene of bacillus subtilis BS04 and application thereof
A technology of Bacillus subtilis and uric acid oxidase, which is applied in oxidoreductase, genetic engineering, plant genetic improvement, etc., can solve the problems of poor thermal stability, low enzyme activity, and limited application, and achieve good heat resistance and efficient degradation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Cloning of Example 1 Bacillus subtilis BS04 urate oxidase gene
[0030] Genomic DNA of Bacillus subtilis BS04 was extracted by CTAB method, and a pair of specific primers Uri-F (5'-ATGTTCACAATGGATGACCTG-3') and Uri-R (5' -GGCTTTCAGGCTC-3'), with genomic DNA as template, Uri-F and Uri-R as primers, PCR amplification is carried out to obtain the uric acid oxidase gene of Bacillus subtilis BS04.
[0031] The PCR reaction program is PCR reaction system: ddH 2O (16.2 μL), 10×buffer (2.5 μL), dNTP (2 μL), Uri-F (1 μL), Uri-R (1 μL), DNA (2 μL), TaqE (0.3 μL), total volume 25 μL. PCR reaction program: 94°C 4min; 94°C 30s, 62°C 30s, 72°C 2min 15s, a total of 35 cycles; finally 72°C extension 7min. The PCR amplified product was recovered and purified with Tiangen gel extraction kit, then ligated with pEASY-T1 vector (Takara) and transformed into E. coli DH5α competent cells, and positive clones were selected to extract plasmids for sequencing. Sequencing results show that the...
Embodiment 2
[0032] Example 2 Construction of Bacillus subtilis BS04 urate oxidase gene prokaryotic expression vector
[0033] According to the gene sequence of uric acid oxidase of Bacillus subtilis BS04 and the multiple cloning site of the prokaryotic expression vector PET-28a (+), a pair of specific primers Uri-P-F (5'-CCC GAATTC ATGTTCACAATGGATGACCTG-3') and Uri-P-R (5'-TT GCGGCCGC GGCTTTCAGGCTC-3'), EcoRI and NotI restriction sites were added to the upstream and downstream primers, respectively. Using Bacillus subtilis BS04 genomic DNA as template and Uri-P-F and Uri-P-R as primers, PCR amplification was carried out.
[0034] PCR reaction system: ddH 2 O (16.2 μL), 10×buffer (2.5 μL), dNTP (2 μL), Uri-P-F (1 μL), Uri-P-R (1 μL), DNA (2 μL), TaqE (0.3 μL), total volume 25 μL. PCR reaction program: 94°C 4min; 94°C 30s, 62°C 30s, 72°C 2min 15s, a total of 35 cycles; finally 72°C extension 7min. The PCR product and the prokaryotic expression vector PET-28a(+) were digested with EcoRI...
Embodiment 3
[0035] Induced expression and purification of embodiment 3 recombinant protein
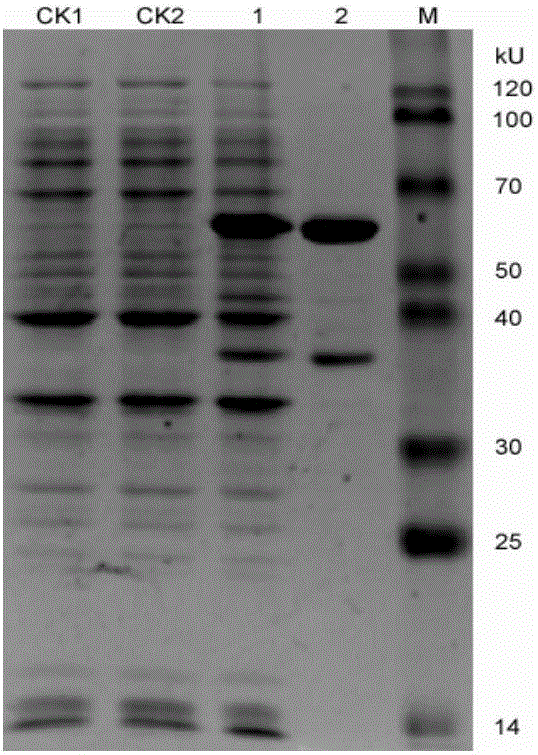
[0036] Take 1 ng of the recombinant plasmid to transform Escherichia coli BL21(DE3), spread it on an ampicillin plate containing 50 μg / mL for overnight culture at 37°C. Positive transformants were picked and cultured in liquid LB medium containing 50 μg / mL ampicillin at 37 °C and 200 r / min until the exponential growth phase, and then IPTG was added to make the final concentration 0.1 mmol / L and cultured for 4 h. The cells were collected by centrifugation and ultrasonically broken, and then 5× loading buffer was added for 12% SDS-PAGE electrophoresis to detect the expression of the recombinant protein. SDS-PAGE electrophoresis was used to detect the expression of the recombinant protein, and the molecular weight of the recombinant protein was about 60kDa.
[0037] Expand the positive recombinant bacteria at a ratio of 1%, induce recombinant expression with IPTG, collect the bacterial cells by cent...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com