Recombinant bacteria for producing salidroside and analogue thereof, and construction method and use
A technology of salidroside and its analogues, which is applied in the field of recombinant bacteria producing salidroside and its analogues and construction, can solve problems such as unfavorable industrial production, achieve industrial production, solve source problems, and reduce production cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
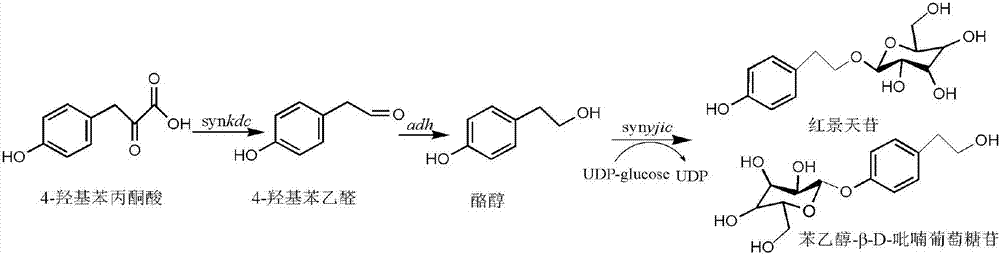
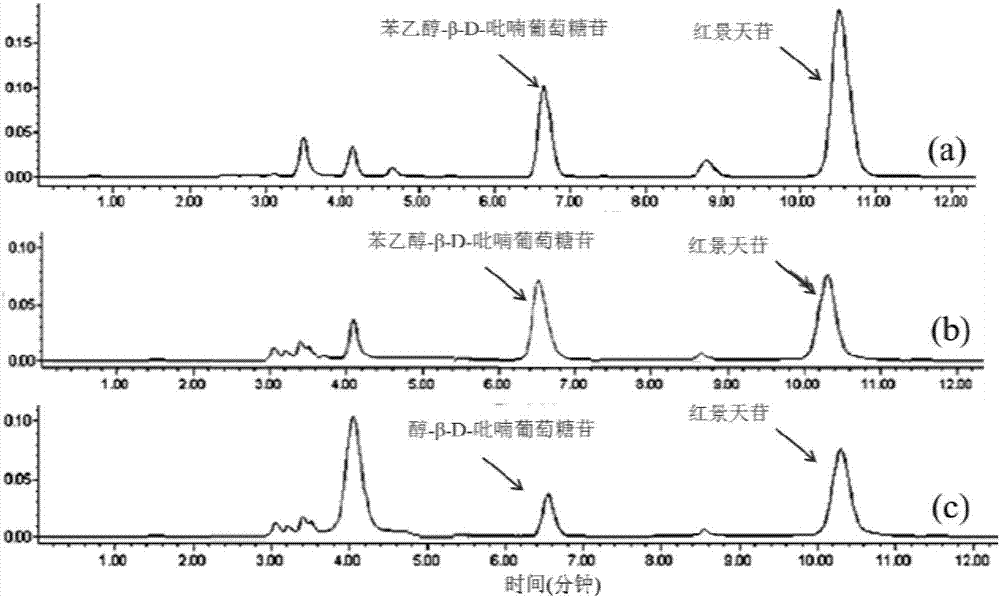
[0042] Example 1 Design of ketone decarboxylase gene synkdc and glycosyltransferase gene synyjic
[0043] Preferably, the Pichia pastoris GS115 keto-decarboxylase gene and the Bacillus licheniformis glycosyltransferase gene, the amino acid sequences of which are respectively shown in SEQ ID No.01 and SEQ ID No.02 in the sequence listing. Use the JCAT online codon optimization software (http: / / www.jcat.de) in combination with the OPTIMIZER online codon optimization tool (http: / / genomes.urv.es / OPTIMIZER / ) to perform codon preference in Escherichia coli Optimizing and designing the full-length synkdc gene and synyjic gene, as shown in the sequence table SEQ ID No.03 and SEQ ID No.04 respectively.
Embodiment 2
[0044] Example 2 λ-red homologous recombination method T7 RNA polymerase was integrated into the chromosome of the chassis strain SyBE-002447.
[0045] The detailed steps of the construction process of the chassis strain integrating T7RNA polymerase into the chromosome of the chassis strain SyBE-002447 are as follows:
[0046] 1. Design the primers T7-RNA F and T7-RNA R sequences respectively as shown in the sequence table SEQ ID NO.07, SEQ ID NO.08 to obtain the T7 RNA polymerase gene from Escherichia coli BL21 (DE3) bacteria, and the T7 RNA polymerase The gene sequence is as shown in the sequence listing SEQ ID NO.05.
[0047] 2. The sequences of primers T7F and T7R, T7-Chl F and T7-Chl R are as shown in the sequence table SEQ ID NO.09, SEQ ID NO.10, SEQ ID NO.11, SEQ ID NO.12, prepared by overlap extension PCR with chlorine Mycin-resistant T7 RNA polymerase fragment.
[0048] 3. Introduce the pKD46 plasmid into the strain SyBE-002447 to obtain SyBE-002447 / pKD46, inoculate...
Embodiment 3
[0051] Example 3 Construction of recombinant expression vector pSynkdc1-yjic
[0052] The synkdc gene fragment obtained by chemical total synthesis was added to the two ends of the fragment by PCR method, respectively, and the EcoRI and PstI restriction sites located downstream of the first promoter of the expression vector pRSFDuet-1 were added, and the FastDigest endonuclease EcoRI was used to Digest with PstI, the reaction system is: 5 μL 10*FD buffer, 2.5 μL EcoRI, 2.5 μL PstI, 30 μL synkdc gene fragment and 10 μL ultrapure water. The reaction conditions are: 37°C, 2h. Using a PCR purification kit, add 250 μL of Bingding Buffer solution to 50 μL of the digested product, mix well, add to the adsorption column, let stand for one minute, centrifuge at 10,000 g for 1 minute, and discard the effluent. Add 650μL Wash Buffer, centrifuge at 10,000g for 1 minute, and discard the effluent. Centrifuge at 10,000g for 2 minutes to remove residual Wash Buffer. Put the adsorption colu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com