Ustilaginoidea virens spore real-time quantification loop-mediated isothermal amplication detection method and kit
A technology of ring-mediated isothermal and rice smut bacteria, applied in the direction of microbial determination/testing, microbial-based methods, biochemical equipment and methods, etc., can solve the problems of complex processing process, difficult prevention and control, and low detection sensitivity. Achieve the effect of optimized reaction temperature, high specificity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
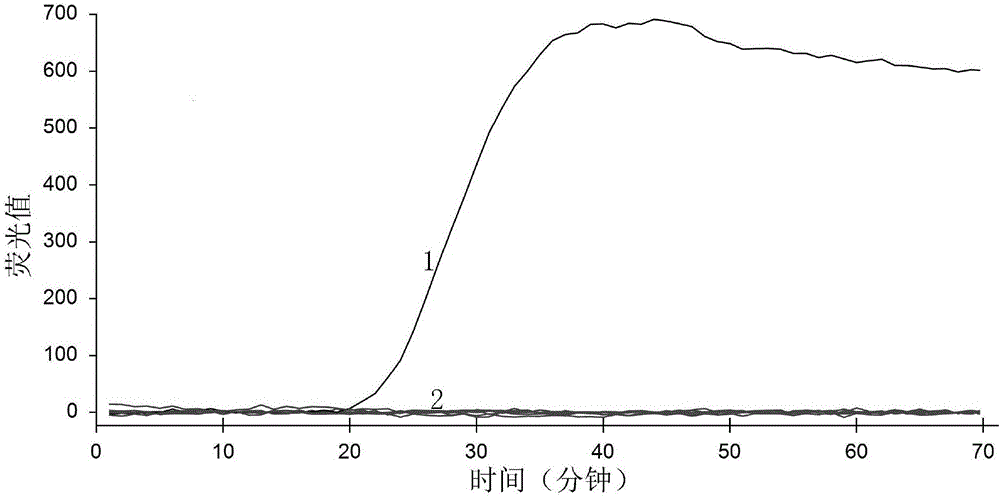
[0069] Embodiment 1, the specificity analysis of primer
[0070] Ustilaginoidea virens was used as a positive control, Fusarium fujikurio, Fusarium graminearum, Penicilliu sp., Pyricularia grisea, Alternaria Alternaria alternate and Rhizoctonia solani were used as negative controls, and double distilled water was used as blank control for testing.
[0071] Use a 20 μL reaction system, which is composed of the following components: 8U / μL Bst DNA polymerase 0.5 μL (4U), 10×ThermoPol Buffer 2.5 μL, 20 μM FIP 2.0 μL, 20 μM BIP 2.0 μL, 10 μM F3 0.5 μL, 10 μM B30 .5 μL, 25 mM MgCl 2Add 4.0 μL, 2.5 μL of 10 mM dNTPs, 3.0 μL of 5M betaine, 1 μL of 10×SYBR Green I, add 1 μL of DNA solution as a template, and make up to 20 μL with double distilled water.
[0072] Experiments were performed with the following primer combinations:
[0073] Upstream outer primer (F3): 5'—CGGCCCTTATACCCCATTC—3';
[0074] Downstream outer primer (B3): 5'—GAGGATCTTCAAGCACCTTGG—3';
[0075] Upstream inner...
Embodiment 2
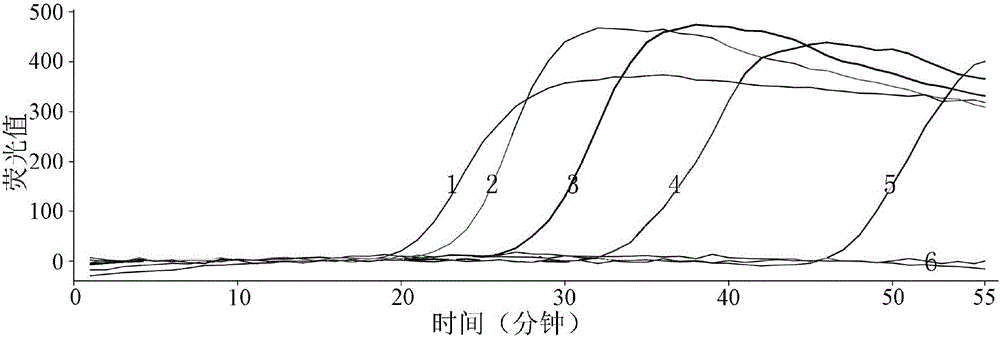
[0099] Embodiment 2, the sensitivity detection of primer
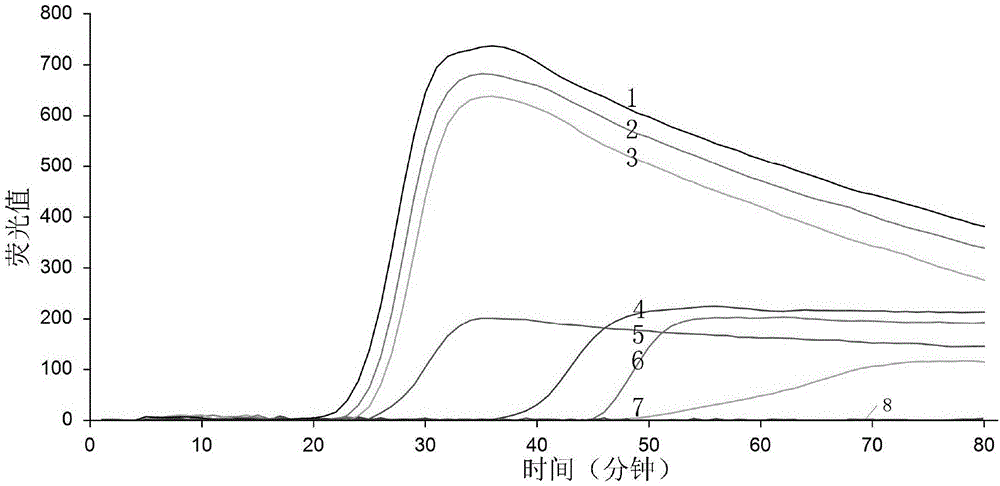
[0100] The DNA of the rice smut fungus (Ustilaginoidea virens) genome was extracted, and the concentration of the DNA solution was measured to be 200 ng / μL by an ultraviolet spectrophotometer. Dilute the DNA solution into 7 concentrations (10ng / μL~1ng / μL) according to a 10-fold gradient. 10ng / μL, 100ng / μL. Configure the following 20μL reaction system: 8U / μL Bst DNA polymerase 0.5μL (4U), 10×ThermoPol Buffer 2.5μL, 20μM FIP 2.0μL, 20μM BIP 2.0μL, 10μM F3 0.5μL, 10μM B3 0.5μL, 25mM MgCl 2 4.0 μL, 2.5 μL of 10 mM dNTPs, 3.0 μL of 5M betaine, 1 μL of 10×SYBR Green I, 1 μL of DNA templates of the above-mentioned concentration gradients (1 μL of double-distilled water was used as blank control), and finally made up to 20 μL with double-distilled water. Experiments were performed with primer combination UV-2. The q-LAMP amplification reaction program was: 64°C, 1min, 60 cycles; 80°C, 10min.
[0101] After the reaction i...
Embodiment 3
[0103] Embodiment 3, the q-LAMP detection of rice false smut fungus spore DNA
[0104] Extraction of the genome of the spore suspension: dilute the spores of the rice smut with 10% SDS to obtain 1.0×10 4 , 2.0×10 3 , 4×10 2 , 8×10 1 , 1.6×10 1 , 3.2×10 0 , 5.4×10 -1 per ml spore suspension. Put 1ml of spore suspension into 2.0ml centrifuge tube, add 0.4g pickled beads, place in Fsat-prep instrument, rotate at 6m / s for 40s, repeat three times, place on ice for 2min. Take 1 μl of the cooled lysate directly as a DNA template for LAMP detection. Experiments were performed with the following primer combinations:
[0105] Upstream outer primer (F3-1): 5'—CGGCCCTTATACCCCATTC—3';
[0106] Downstream outer primer (B3-1): 5'—GAGGATCTTCAAGCACCTTGG—3';
[0107] Upstream inner primer (FIP): 5'—TCTACCCCGCAACTCAGCCCGCGGTTCGTCCGTTTTCG—3';
[0108] Downstream internal primer (BIP): 5'—AGGAGGGGAACTGACACAGGGATTGGCCATGCGTCCATC—3'.
[0109] A 20 μL reaction system was prepared by propo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com