Pseudomonas and separating method and application thereof
A technology of Pseudomonas and Pseudomonas strains, which is applied in the field of microorganisms, can solve the problem that the streak bacteria do not show resistance, and achieve a significant antagonistic effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1, the acquisition of Pseudomonas 1257
[0040] 1. Soil source
[0041] Rhizosphere Soil of Chinese Cabbage in Vegetable Field in Fengxian District, Shanghai
[0042] 2. Screening of bacterial strains
[0043] (1) Soil sample collection
[0044] Zig-shaped 5-point sampling method for soil sample collection: collect 200g soil samples at each point, mix the soil samples at 5 points taken according to the Z-shape evenly, take 200g by quartering method and put them into a sterilization bag as a soil sample . Three soil samples were collected from each plot as replicates. Record the time, place and type of sampling. The collected soil samples were stored in a refrigerator at 4°C for use in bacterial isolation.
[0045] (2) Isolation of bacteria
[0046] Plate dilution method: Weigh 10g of each soil sample into a sterile conical conical flask, add 90mL of sterile water, then place at 200rpm, shake in a shaker at 28°C for 15min, take out the conical flask and...
Embodiment 2
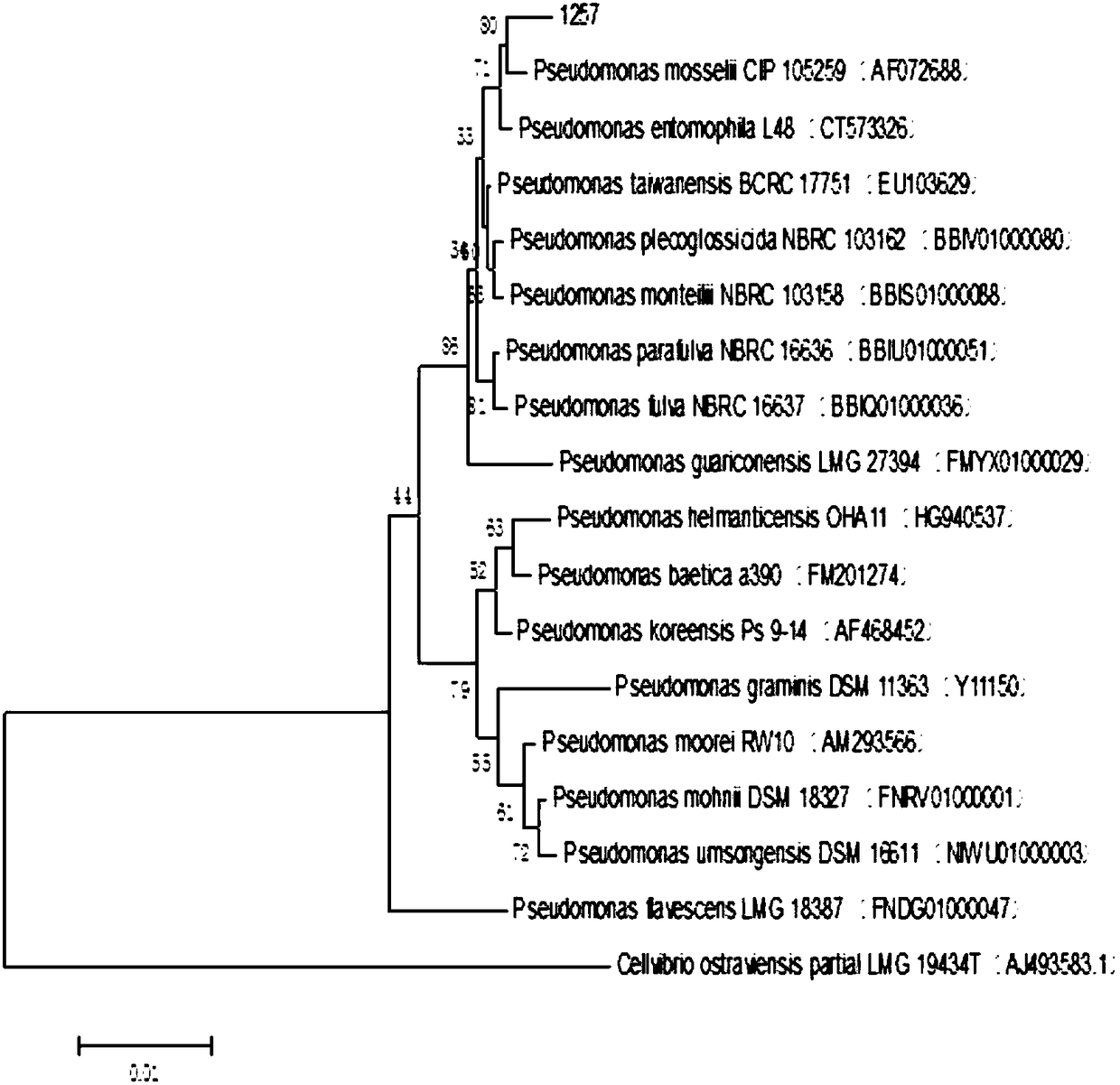
[0052] Embodiment 2, the identification of the 16S rRNA gene of Pseudomonas 1257
[0053] Genomic DNA of strain 1257 was extracted using primer: 27F5'-AGAGTT TGA TCCTGGCTCAG-3' and 1492R5'-TACGGCTACCTTGTTACGACTT-3'; using the extracted DNA as a template, perform PCR amplification to obtain the target fragment. The PCR reaction system is:
[0054]
[0055] The reaction mixture was mixed gently and then briefly centrifuged to perform PCR amplification. The PCR reaction conditions are:
[0056] 94°C for 3min,
[0057] 94°C 30sec, 56°C 30sec, 72°C 90sec, 30 cycles,
[0058] 72℃8min,
[0059] 25°C Forever.
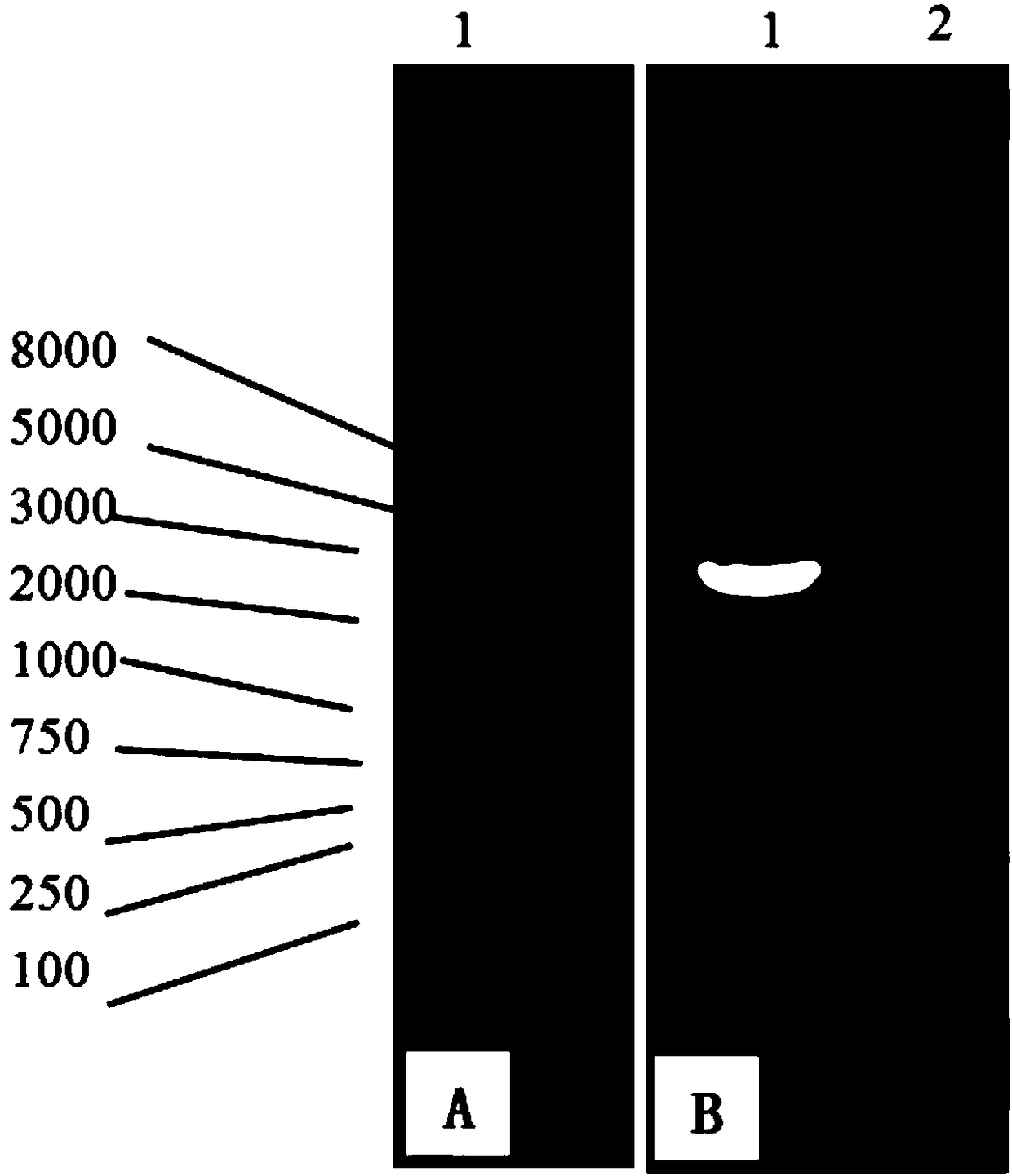
[0060] Take 5 μL of the above-mentioned PCR amplification product, and use 1% agarose gel for electrophoresis detection (see image 3 ), the PCR stock solution was sent to Platinum Biotechnology (Shanghai) Co., Ltd. for sequencing, and its base sequence is shown in SEQ ID No.1.
[0061] The sequencing results were analyzed using DNA Star and compared with BLAST on t...
Embodiment 3、1257
[0063] Physiological and biochemical identification of embodiment 3, 1257
[0064] The physiological and biochemical characteristics of Pseudomonas 1257 of the present invention are: it can produce fluorescent pigment, can secrete arginine dihydrolase and protease, and can assimilate glucose, mannose, mannitol, gluconate, capric acid, apple Acid, citric acid, phenylacetic acid, see Table 1. Can utilize 62 kinds of carbon sources such as Tween 40, L-arabinose, D-fructose, α-D-glucose, etc. It cannot hydrolyze indole, escin, and β-galactosidase, and cannot reduce nitric acid to nitrite, see Table 2.
[0065] Table 1 Physiological and biochemical characteristics of 1257 - enzyme activity, carbon source assimilation
[0066]
[0067] +: Positive reaction; -: Negative reaction;
[0068] Table 2 Physiological and biochemical characteristics of 1257—acid production using carbon source
[0069]
[0070]
[0071]
[0072] +: Positive reaction; -: Negative reaction; W: W...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com