7beta-hydroxyl sterol dehydrogenase mutant and application thereof to preparation of ursodeoxycholic acid
A technology of hydroxysterol dehydrogenase and mutant, which is applied to 7β-hydroxysterol dehydrogenase mutant and its application field in the preparation of ursodeoxycholic acid, can solve the problem of high price, high application cost of coenzyme and catalyst preparation process Cumbersome and other problems, to achieve the effect of improving thermal stability and efficient utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
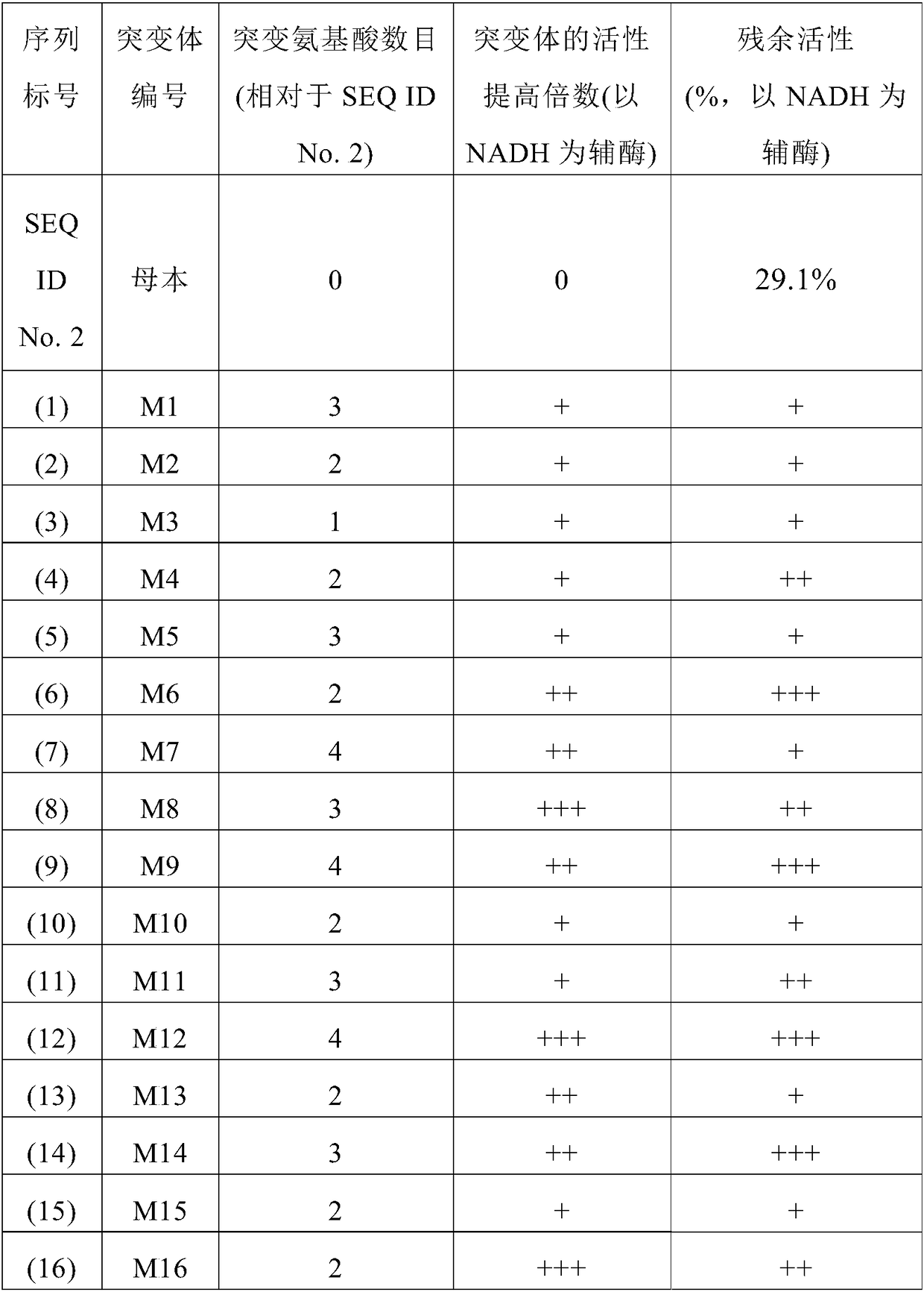
[0071] Example 1 Site-directed mutation of 7β-HSDH
[0072] Through Uniprot, NCBI BLAST and spatial structure modeling, the amino acid residues around the binding site of the coenzyme NADPH include: amino acid, 18-position glutamic acid, 22-position lysine, 39-position glycine, 44-position lysine and 67-position phenylalanine, etc. Using site-directed mutagenesis technology, the amino acid residues at these sites were subjected to site-directed mutation. Through screening, it was found that the 17th threonine was replaced by alanine (T17A), and the 18th glutamic acid was replaced by threonine (E18T). ), the 22nd lysine was replaced with aspartic acid (K22D), the 39th glycine was replaced with aspartic acid (G39D), the 44th lysine was replaced with glycine (K44G), the 67th phenylpropanoid Mutants such as amino acid replaced by alanine (F67A) greatly increased the activity of NADH, and correspondingly, the specific activity of NADPH decreased significantly.
[0073] The 7β-HSD...
Embodiment 2
[0074] Example 2 Construction of 7β-HSDH mutants
[0075] On the basis of the mutant described in Example 1, the error-prone PCR technique was used for random mutation to further improve the activity of the enzyme.
[0076] According to the open reading frame of 7β-HSDH, design upstream and downstream primers as follows:
[0077] Upstream primer, as shown in SEQ ID No.3:
[0078] CCG GAATTC ATGAATCTGCGTGAAAAATAC
[0079] Downstream primer, as shown in SEQ ID No.4:
[0080] CCG CTCGAG TTAATTGTTGCTATAGAAGC
[0081] Wherein, the sequence indicated by the underline of the upstream primer is the restriction site of EcoR I, and the sequence indicated by the underline of the downstream primer is the restriction site of Xho I.
[0082] Using pET28a-7β-HSDH as a template, error-prone PCR was performed with rTaq DNA polymerase to construct a random mutation library. PCR system (50 μL): rTaq DNA polymerase 0.5 μl, 10×PCR buffer (Mg 2+ Plus) 5.0 μl, dNTP Mixture (2.0 mM each) 4...
Embodiment 3
[0105] Example 3 Recombinant E.coli BL21(DE3) / pET28a-7β-HSDH M12 expression and activity assay
[0106] The recombinant Escherichia coli E.coli BL21(DE3) / pET28a-7β-HSDH of the mutant M12 obtained in Example 2 M12 Inoculated into the LB medium containing 50 μg / ml kanamycin, cultured in a shaker at 37°C for 12 hours, and then inserted into 100ml LB medium (containing 50 μg / ml Kanamycin) in a 500ml Erlenmeyer flask, placed at 37°C, 180rpm shaker culture, when the OD of the culture solution 600 When it reached 0.6, IPTG with a final concentration of 0.2mmol / L was added as an inducer and induced at 16°C for 24h. The culture solution was centrifuged at 8000×g for 10 min, the cells were collected, and washed twice with saline to obtain resting cells. Suspend the cells obtained in 100ml of culture medium in 10ml of potassium phosphate buffer (100mM, pH 8.0), and perform the following ultrasonic disruption in an ice-water bath: 400W power, working for 4s, intermittent for 6s, for 99...
PUM
| Property | Measurement | Unit |
|---|---|---|
| specific rotation | aaaaa | aaaaa |
| purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com