Saccharomyces cerevisiae S8-H with high yield of xylanase and application
A technology of Saccharomyces cerevisiae and Saccharomyces cerevisiae strains, applied in the field of microbial engineering, can solve the problems of lack of high-yielding strains and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Overlap extension PCR method to construct PαXC expression cassette
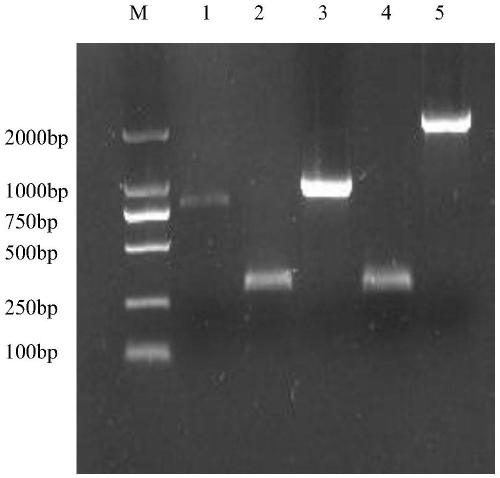
[0027] Aspergillus niger CICC2462 strain (China Industrial Microorganism Culture Collection Center) was subjected to PDA (composition: 200g potato boiled liquid, 20g glucose, 1.5g MgSO 4 ·7H 2 O, 1g KH 2 PO 4 , 20g Agar, deionized water constant volume to 1L) after plate activation, prepare seed liquid, seed liquid is inoculated in liquid fermentation medium (composition is: CaCl 2 5g, KH 2 PO 4 26g, FeSO 4· 7H 2 O 0.16g, yeast extract 10g, bran 20g, MgSO 4· 7H 2 O 10g, Tween-80 5ml, pH 5.0, distilled water 1000mL) 30°C, 150rpm, cultured for 72h, collected bacteria, and extracted RNA with AXYGEN total RNA extraction kit. Use the Taraka one-step reverse transcription kit to synthesize cDNA to amplify the xylanase xynB gene, the upstream and downstream primers xynB-F and xynB-R nucleotide coding sequences such as SEQ ID NO: 1 and SEQ ID NO: 2, Using S.Cerevisiae INVSc1 (purchased fr...
Embodiment 2
[0032] Example 2 Construction of S.Cerevisiae INVSc1-pYES2-PαXC-rDNA engineering bacteria
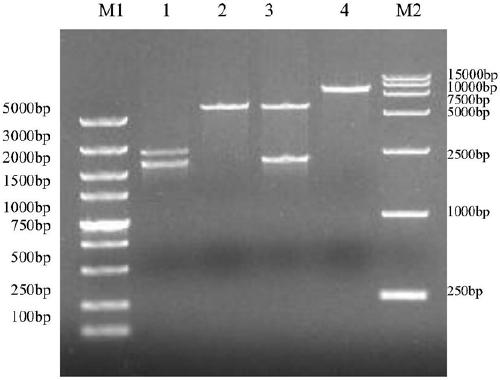
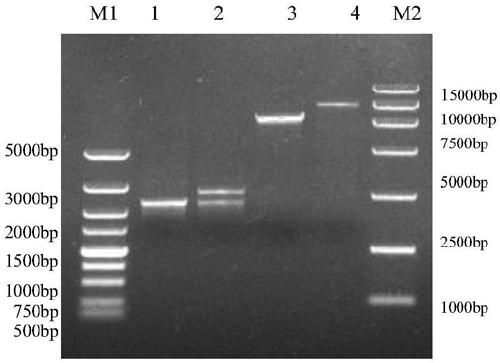
[0033] Using S.Cerevisiae INVSc1 genomic DNA as a template, amplify the core sequence of 2300bp (Genebank accession number BK006945.2) in the rDNA unit, the nucleotide sequence is as SEQ ID NO: 10, the upstream and downstream primers rDNA-F and rDNA-R core Nucleotide coding sequences such as SEQ ID NO: 11 and SEQ ID NO: 12, pYES2-PαXC and pMD19-T-rDNA were single-digested with SnaBI respectively, the target fragments were recovered, ligated with T4 ligase, and sequenced to obtain pYES2-PαXC and pMD19-T-rDNA. PαXC-rDNA expression vector such as image 3 shown. After the pYES2-PαXC-rDNA vector was linearized by SphI at the rDNA position, the LiAc / ssDNA method was used to transform the competent S. Cerevisiae INVSc1, and the transformed system was spread on the auxotrophic SC-U medium without uracil ( Purchased from Japan Clonetech Company), xylanase activity screening medium adopts xyla...
Embodiment 3
[0034] Example 3 Microdrop digital PCR identification of transformant copy number
[0035] For the obtained positive transformants, inoculate in YPD medium containing 20g / L peptone, 10g / L yeast powder and 10g / L glucose, the culture condition is 30°C, collect the bacteria after 48 hours of culture, and use Omega yeast genome extraction reagent The cassette extracts DNA, using the DNA as a template, and the ATC1 gene as an internal reference gene sequence (885bp, Genebank accession number CP020126.1) nucleotide coding sequence such as SEQ ID NO:13. XynB gene copy number identification using BIO-RAD QX200 digital droplet PCR. The nucleotide coding sequences of ATC1 upstream and downstream primers ATC1-F1 and ATC1-R1 are as SEQ ID NO: 14 and SEQ ID NO: 15, probe: 5'-HEX-CATCTTCGTTAGCTTCATCCGACGCTA-BHQ-3'; upstream primer xynB-F2 And the nucleotide coding sequence of the downstream primer xynB-R2 such as SEQ ID NO: 16 and SEQ ID NO: 17, probe: 5'-FAM-CCTGGTCAACTTTGCCCAGTCTAACAA-BH...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com