Halophilic denitrifying bacterium YL5-2 and application thereof
A denitrifying bacteria and denitrifying technology, applied in the halophilic denitrifying bacteria YL5-2 and its application field, can solve the problem of less halophilic denitrifying bacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Isolation and preservation of the halophilic denitrifying bacteria
[0027] The halophilic denitrifying bacteria YL5-2 was isolated from the sedimentary soil of Golmucharhan Salt Lake (36°51′N, 94°95′E) in Qinghai Province, China. The water of Chaerhan Salt Lake is saturated or close to saturated salt concentration all year round.
[0028] Prepare LB liquid medium with NaCl concentration of 20%, add 250 mg / L glycerol, 250 mg / L glucose, and 50 mg / L methanol, and enrich and cultivate the depositional soil of Chaerhan Salt Lake for 48 hours at 30°C. The strains in the enriched culture medium were isolated by using YL solid medium. 1L medium contains the following components: glucose: 0.6g, trisodium citrate 0.5g, glycerol 2mL, yeast extract 0.8g, peptone 1.6g, dipotassium hydrogen phosphate 0.35g, potassium dihydrogen phosphate 0.1g, ammonium sulfate 0.25g, ammonium chloride 0.25g, MgSO 4 0.5g, CaCl 2 0.1g, NaCl 180g; trace element SL-4 10mL, pH 7.0-7.2; ag...
Embodiment 2
[0030] Example 2 Analysis of 16S rRNA sequence and whole gene sequence of halophilic denitrifying bacteria YL5-2
[0031] Strain YL5-2 genomic DNA was extracted using TaKaRa kit (TaKaRa MiniBEST Bacteria Genomic DNA Extraction 68Kit Ver.3.0).
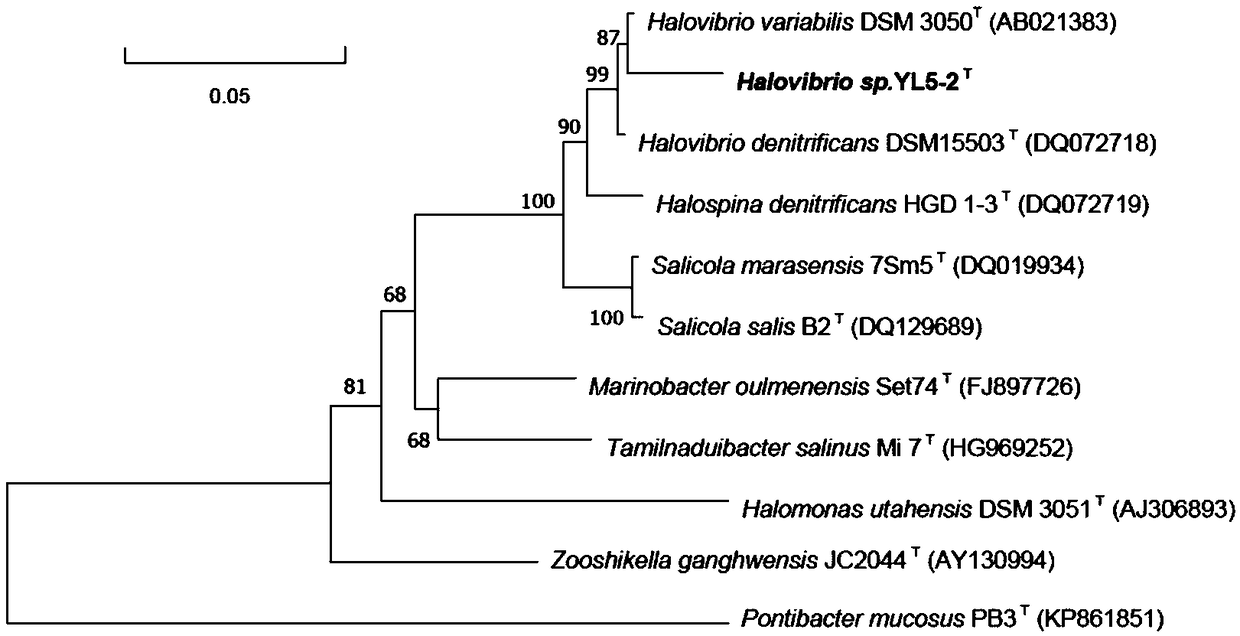
[0032] 16S rRNA was amplified using universal bacterial primers 27F (5’-AGAGTTTGATCMTGGCTCA G-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’). PCR sequencing was entrusted to Shanghai Sangon Biotechnology Co., Ltd. The complete 16S rRNA sequence of strain YL5-2 is 1518bp, as shown in SEQ ID NO.1, and the GenBank accession number is MF782425.
[0033] The whole genome of strain YL5-2 was sequenced using the IlluminaMiSeq 2000 high-throughput sequencing platform of Shanghai Passino Biotechnology Co., Ltd. The original sequencing data was filtered and corrected using PRINSEQ (version number v 0.20.4) software, and then SOAP denovo software (version number v1.05) software with default parameters was used for genome base pairing, and then Check...
Embodiment 3
[0038] Example 3 Identification of phenotypic and physiological and biochemical characteristics of halophilic denitrifying bacteria YL5-2
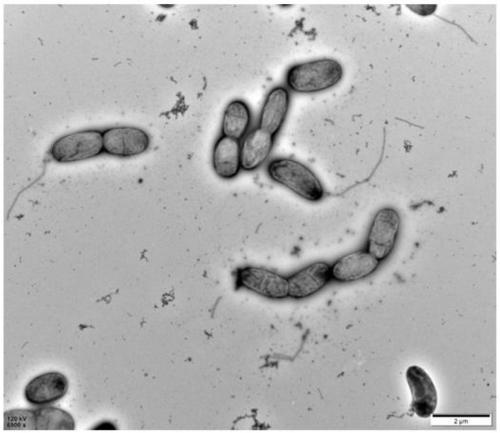
[0039] Gram staining properties were tested using the BD Gram staining kit.
[0040] Cell motility was determined using half MA medium (0.5% agar, w / v).
[0041] Cell morphology was detected by transmission electron microscopy (TEM) analysis. That is, cells were picked from exponentially growing culture medium, stained with 0.5% uranyl acetate, and photographed under a microscope (Tecnai Spirit, FEI, Hillsboro, OR, USA).
[0042] Oxidase activity using the oxidase kit (bioMérieux), by adding 3.0% H 2 o 2 Catalase activity was determined by pouring the solution onto bacterial colonies and observing the generation of bubbles.
[0043] The temperature growth conditions were carried out on YL liquid agar medium, the temperatures were 4, 10, 15, 20, 25, 30, 33, 37, 40, 45 and 50 ° C, and the pH was constant at 7.5, and the strain YL5- 2 T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com