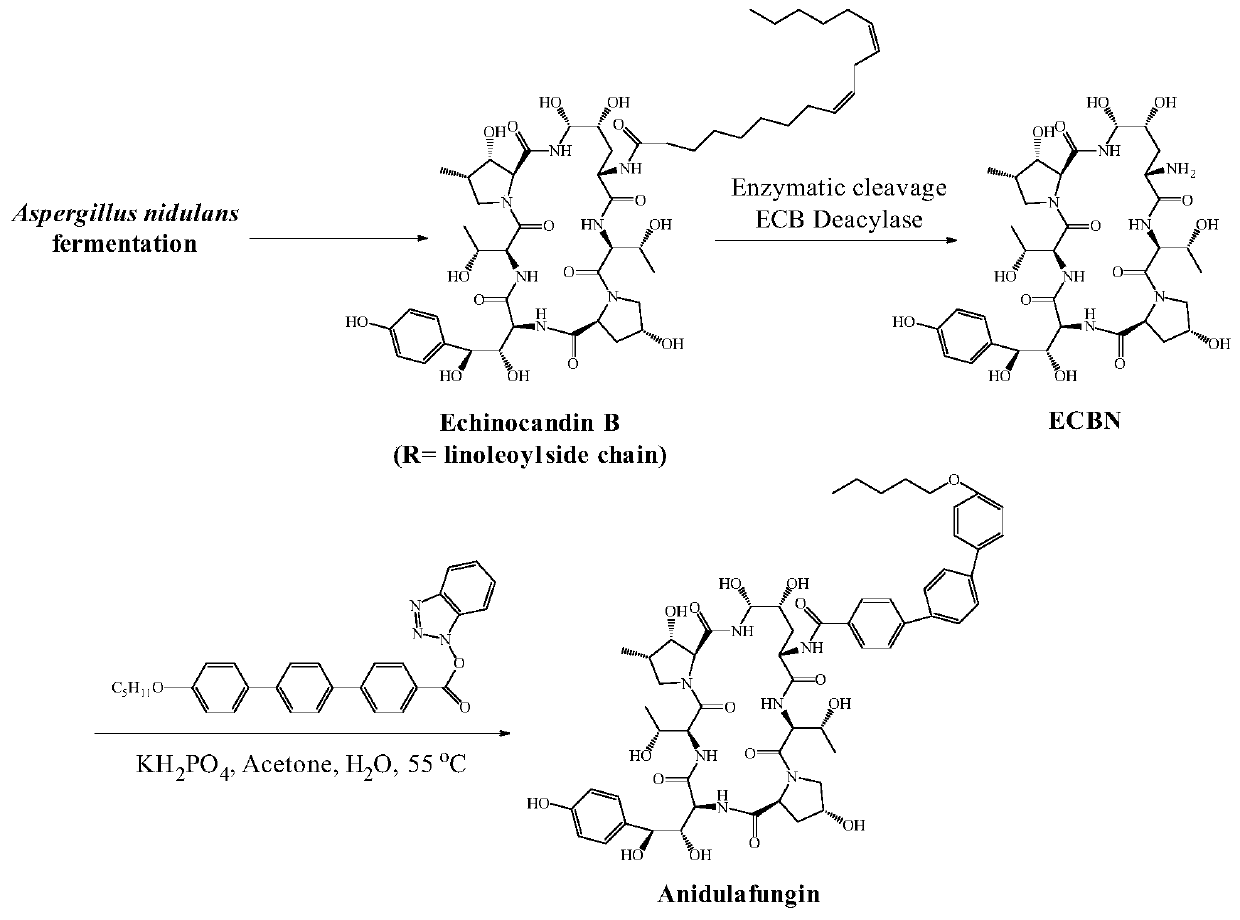
Recombinant ECB (echinocandin B) deacylase mutant and application
A technology of deacylase and echinocandin, applied in the direction of recombinant DNA technology, bacteria, hydrolase, etc., can solve the problem of low enzyme activity and achieve the effect of improving catalytic performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Homology Modeling and Molecular Docking
[0040] In the SWISS-MODEL database (https: / / www.swissmodel.expasy.org / ), the amino acid sequence of the deacylase AuEBDA (shown in SEQ ID No.2, the nucleotide sequence of the coding gene auadea is SEQ ID No. 1) for comparison. The model protein with the highest degree of homology is the antibiotic acyltransferase MacQ (PDB ID: 5C9i_1) derived from Acidovoraxsp.MR-S7, whose amino acid sequence has a homology of 40.77% with the starting deacylase AuEBDA.
[0041] Modeler 9.14 software was used for program coding, and the MacQ protein sequence was used as a template to construct a three-dimensional space model of the AuEBDA protein sequence, and the best model was obtained. Subsequently, the ChemDraw software was used to draw and convert the three-dimensional model of the substrate ECB, and the AutoDock 4.2 software was used for molecular docking. Using Pymol1.6.0.0 to analyze the three-dimensional structure relationsh...
Embodiment 2
[0044] Embodiment 2: Construction of Escherichia coli cloning vector by PCR method
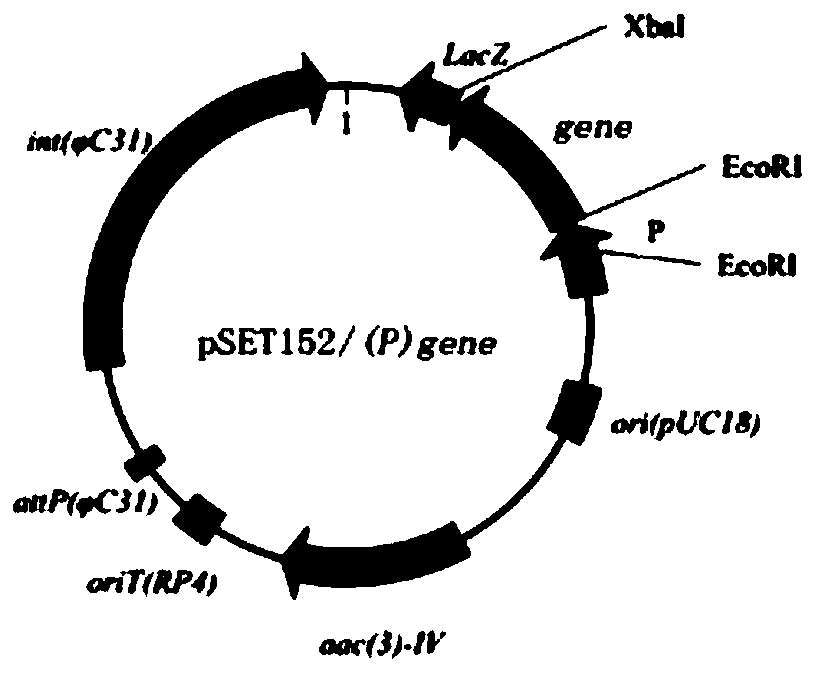
[0045] 1. pSET152-auebda plasmid
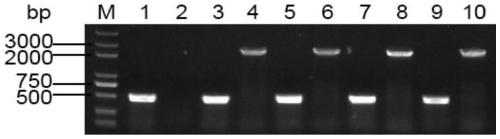
[0046] Actinoplanes utahensis ZJB-08196 (Wang Y J, Liu L L, Feng Z H, et al. Optimization of media composition and culture conditions foracarbose production by Actinoplanes utahensis ZJB-08196[J].World Journal of Microbiology&Biotechnology, 2011, 27 (12): 2759-2766.) Carry out streak activation on the synthetic No. V solid medium without resistance, pick single bacterium colony in the seed medium without resistance, 28 degrees Celsius, 200 revs per minute and cultivate 3 On the next day, the cells of Actinomyces uthaensis ZJB-08196 were obtained, and the genome was extracted. Using the extracted genome as an amplification template, EcoR I and Xba I restriction sites were added to both ends of the gene sequence, and auebda-F and auebda-R were used as primers for PCR amplification (the amplification system and conditions are shown in Table 1 and Table 2), the...
Embodiment 3
[0072] Embodiment 3: Streptomyces lividans expression vector construction
[0073] 1. The plasmids in Example 2 are transformed into E.coli ET12567 / pUZ8002 respectively, and single colonies of positive clones are picked in 10 milliliters of LB medium (containing 50 micrograms per milliliter of kanamycin, 25 micrograms per milliliter of chloramphenicol Incubate overnight at 37°C and 200 rpm in a test tube with 25 micrograms per milliliter of prunamycin. Overnight bacterial solution was transferred to 50 milliliters of LB liquid medium containing corresponding antibiotics with an inoculum of 1% in volume concentration, cultivated at 37 degrees Celsius and 200 rpm for 6 to 8 hours, and the OD 600 Reach 0.4 to 0.6. Wash twice with an equal volume of sterile LB medium, then suspend with 0.5 ml of fresh sterile LB medium, which is the E.coli suspension, and measure the concentration of E.coli with a hemocytometer.
[0074] 2. Prepare the spore suspension of Streptomyces lividans. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com