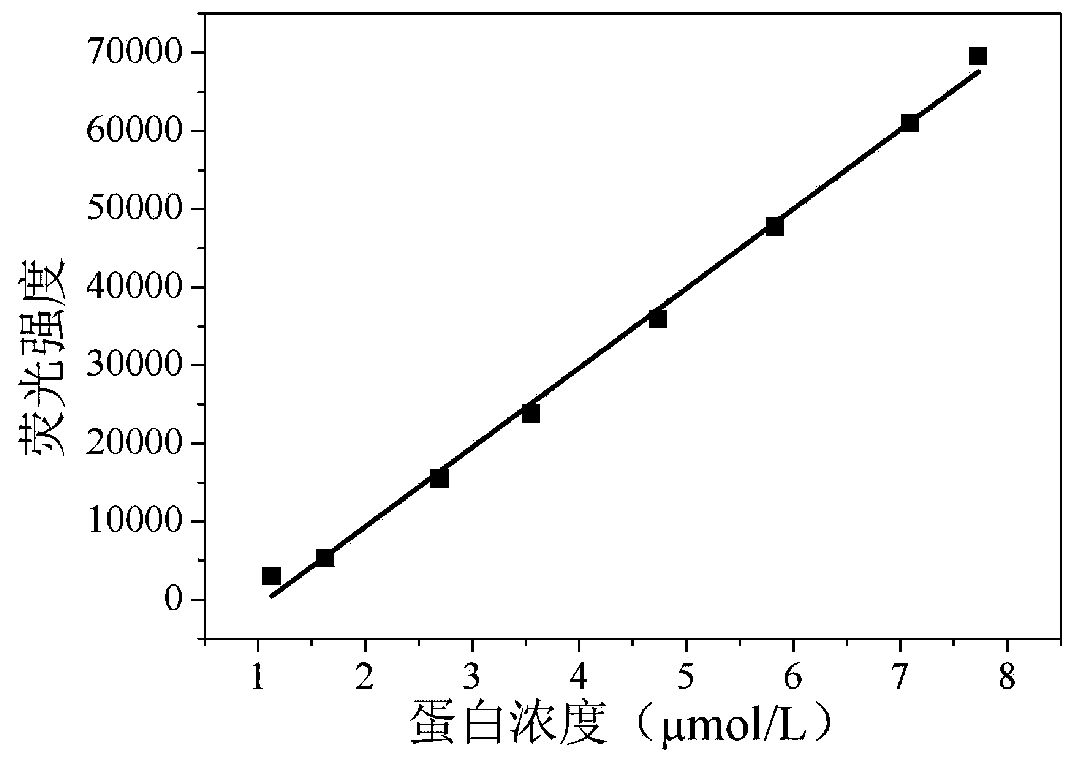
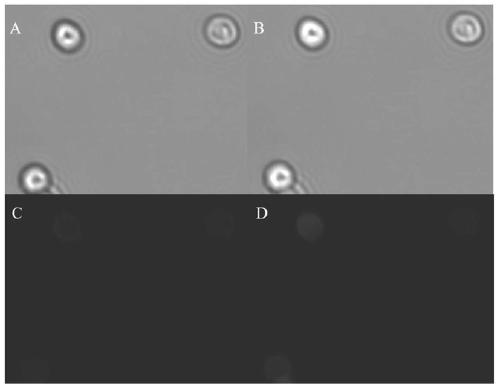
Pichia pastoris surface co-display system based on cellulosome as well as construction method and application of pichia pastoris surface co-display system
A technology of surface co-display and Pichia pastoris, applied in the fields of application, chemical instruments and methods, botany equipment and methods, etc., can solve the problems of difficult foreign protein display efficiency quantification, low amino acid sequence homology, etc., and achieve improvement The effect of display ability, improvement of display ability, and cost saving of preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Embodiment 1: Design and synthesis of MixA2 gene:
[0081] The genome DNA sequences of C.thermocellum and C.cellulolyticum were searched in the NCBI database respectively. The accession numbers of these two microorganisms in the NCBI database are: CP000568 and NC_011898. The coding DNA sequences of the respective cohesin modules were confirmed in these genomes, and then these coding sequences were electronically spliced by DNAMAN software to form preliminary mini-scaffold protein genes. According to the codon usage frequency database information Codon UsageDatabase, the coding MixA2 was optimized according to the preferred codons of Pichia pastoris. Under the premise of not changing the amino acid sequence, the JCat codon optimization software was used to select the optimal codon, and at the same time, fine-tuning was performed with reference to the secondary structure of the mRNA to make the activation energy as high as possible, so that the mRNA was easier to expres...
Embodiment 2
[0082] Example 2: Construction of recombinant plasmid pPICZαA-egfp-mixa2-sed1:
[0083] Mini-scaffold protein MixA2 gene primer: upstream primer P1: 5'-ggaattc catatg gttcgtataaaagttgacacggtc-3' (with Nde I restriction site); downstream primer P2: 5'-aaggaaaaaa gcggccgc acctctacttggtgt-3' (with NotI restriction site).
[0084] Mutated green fluorescent protein gene egfp primer: upstream primer P3: 5'-ccg gaattc agcaagggcgaagagctgttcac-3' (contains EcoR I restriction site); downstream primer P4: 5'-ggattc catatg aagctttacagttcatccatgccgagagt-3' (with Nde I restriction site).
[0085] Sed1 primer of Saccharomyces cerevisiae cell wall protein gene: upstream primer P5: 5'-aaggaaaaaa gcggccgc caattttccaacagtacatctgcttc-3' (with Not I restriction site); downstream primer P6: 5'-catg tctaga ttataagaataacatagcaacaccagccaaac-3' (containing Xba I restriction site).
[0086] PCR reaction system: 1 μL MixA2 synthetic sequence, 1 μL upstream primer P1, 1 μL downstream primer P2...
Embodiment 3
[0090] Example 3: Transfer of recombinant plasmid pPICZαA-egfp-mixa2-sed1 into Pichia pastoris
[0091] The recombinant plasmid vector pPICZαA-egfp-mixa3-sed1 was linearized by PCR with ultra-fidelity DNA polymerase to improve the linearization effect and increase the transformation efficiency of yeast cells. The sequences of the linearization primers are as follows: upstream primer P7: 5'-gagctcgctcattccaattcctt-3', downstream primer P8: 5'-caatcaagcccaataactgggctg-3'.
[0092] Transform into Pichia pastoris KM71H by electric shock, select a single colony on the YPDS plate of Zeocin antibiotic (100 μg / mL), extract the genome as a template, and perform PCR amplification on the target DNA with specific primers, and identify positive according to the size of the PCR band Turn.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com