A high-efficiency polypeptide-polypeptide coupling system and method based on disordered protein coupling enzymes
A polypeptide coupling and protein technology, which is applied to the coupling or cyclization of proteins. The polypeptide-polypeptide coupling system based on disordered protein coupling enzymes can solve the problem of low coupling efficiency of ligases and achieve functional The effect of stability, high reaction efficiency and high purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Transformation and expression of fusion protein target gene
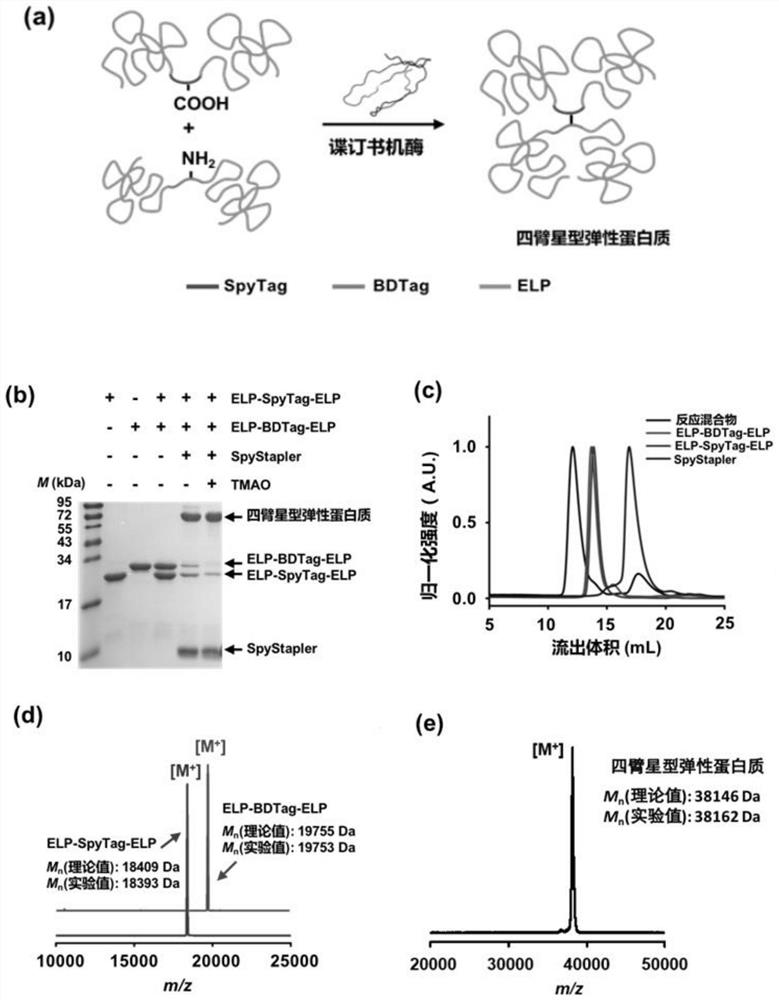
[0032] Plasmids of pQE-80 L ELP-SpyTag-ELP and pQE-80 L ELP-BDTag-ELP were constructed, and after being confirmed by sequencing, they were transferred into BL21 (DE3) competent cells and plated, and left overnight in the incubator for 37 o C culture. Pick out the monoclonal colony on the plate to the LB medium containing 0.10 mg / mL ampicillin sodium, in a shaking incubator for 37 o C was incubated overnight. The overnight culture medium was added to 1 L LB medium containing 0.10 mg / mL ampicillin sodium at a ratio of 1:100, at 37 o C shaking culture to OD 600 For 0.4-0.6, add isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM to induce E. coli to express proteins. Shaking incubator temperature changed to 30 o C culture.
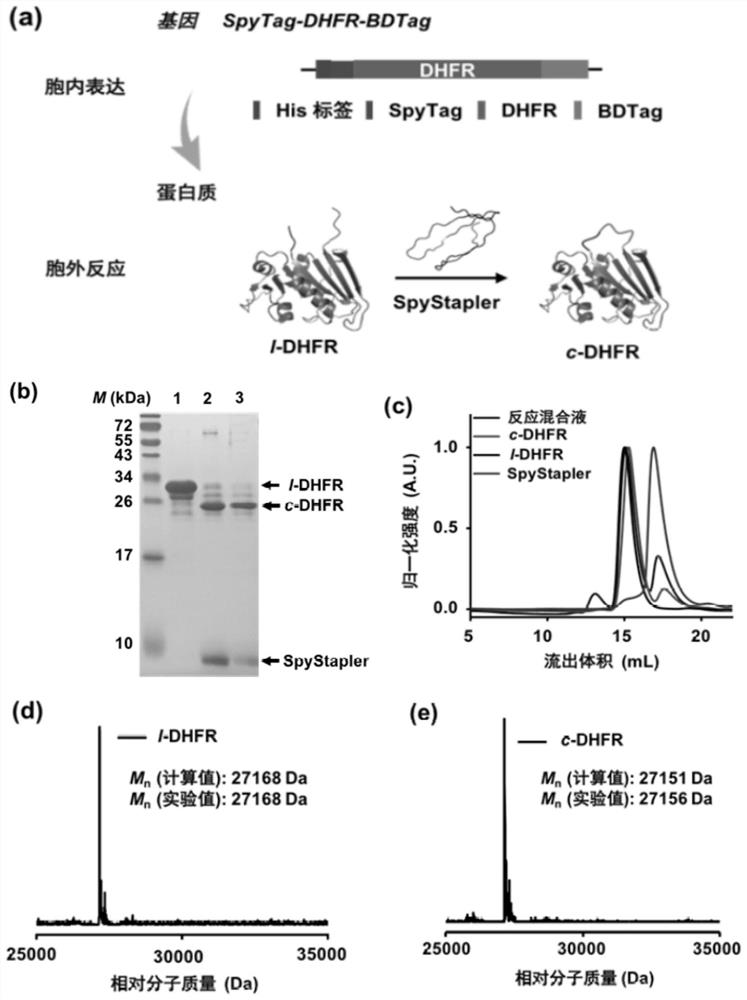
[0033] The plasmid of pACYCDuet-1 SpyTag-DHFR-BDTag (MCS1)-SpyStapler (MCS2) was constructed, and after being confirmed by sequencing, it was transfe...
Embodiment 2
[0034] Embodiment 2: the purification of fusion protein
[0035] After the expression was finished, the cells were collected by high-speed centrifugation at 4000 g for 20 minutes, and the supernatant was discarded. For the protein Spy Stapler enzyme (SpyStapler), spy stapler enzyme mutant (SpyStapler-EQ) and fusion protein SpyTag-DHFR-BDTag purified under natural conditions, use non-denaturing buffer A (20 mM sodium dihydrogen phosphate , 300 mM NaCl, 10 mM imidazole, pH = 8.0) resuspended. Resuspend at 4 o C Sonicate for 20 minutes in an ice-water bath (working for 5 seconds, interval of 5 seconds, intensity 40%). In; 4 o Under the condition of C, the crushed solution was centrifuged at 20,000 g for 20 minutes with a high-speed floor centrifuge, and the lysed supernatant was reserved. The supernatant was mixed with the equilibrated affinity resin Ni-NTA resin at 4 o C Rotate to mix well for 1 hour. Pour the mixed solution into the empty column, wait for the resin to set...
Embodiment 3
[0037] Example 3: Characterization of Fusion Proteins
[0038] 1 mL of the protein eluate was purified through a gel permeation column Superdex 200 Increase 10 / 300 GL on the AKTA protein purification system (AKTA Avant, GE Healthcare). The mobile phase was PBS, and the flow rate was 0.5 mL / min. By monitoring A 280 Collect peaks of interest.
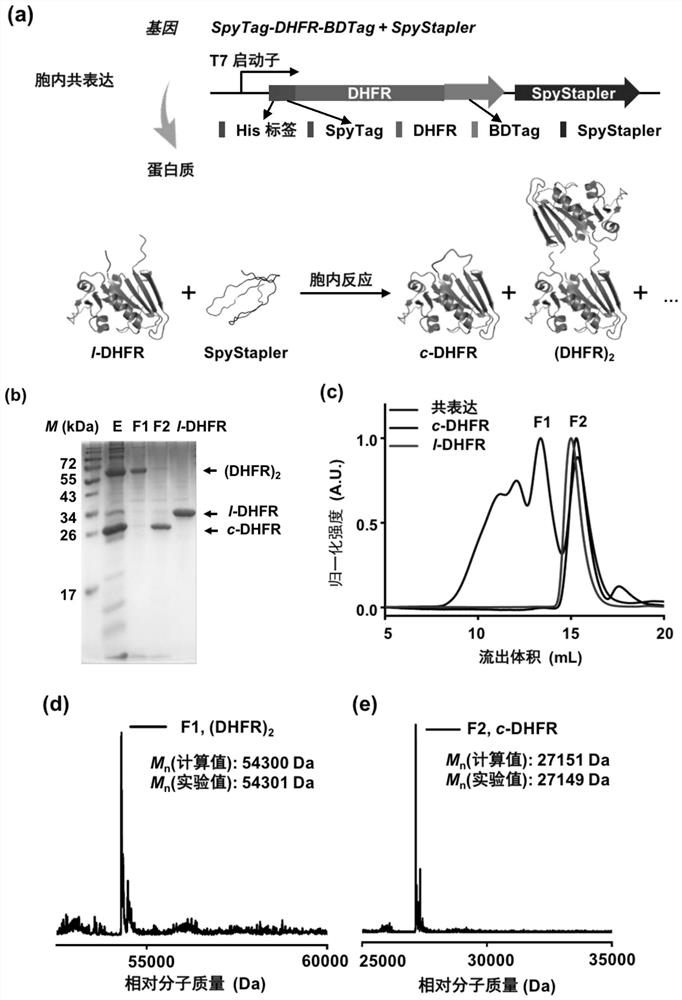
[0039] The molecular weight of the purified product was determined by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF) or high-performance liquid chromatography-electrospray mass spectrometry (LC-MS), and the instrument used was 5800 MALDI-TOF / TOF analyzer (ABSCIEX) . The result is as figure 1 As shown, in vivo SpyStapler-mediated peptide-peptide (BDTag-SpyTag) coupling reaction leads to in situ cyclization of SpyTag-DHFR-BDTag.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com