Ginkgo GbFLSa gene and expression protein and application thereof
A ginkgo and gene technology, applied in the field of ginkgo GbFLSa gene and its expression protein and application, can solve the problems of the function research in plants without key enzyme genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
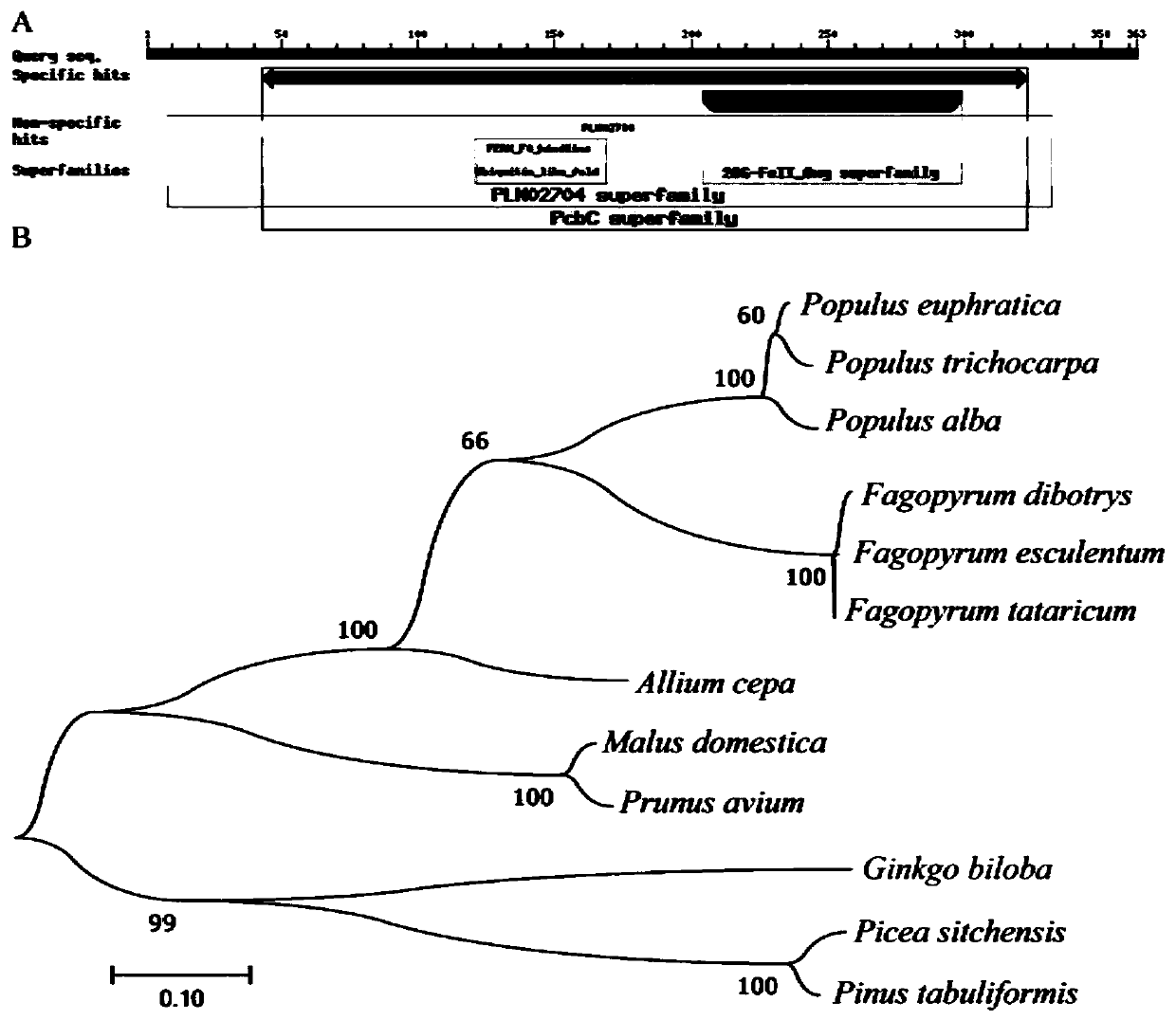
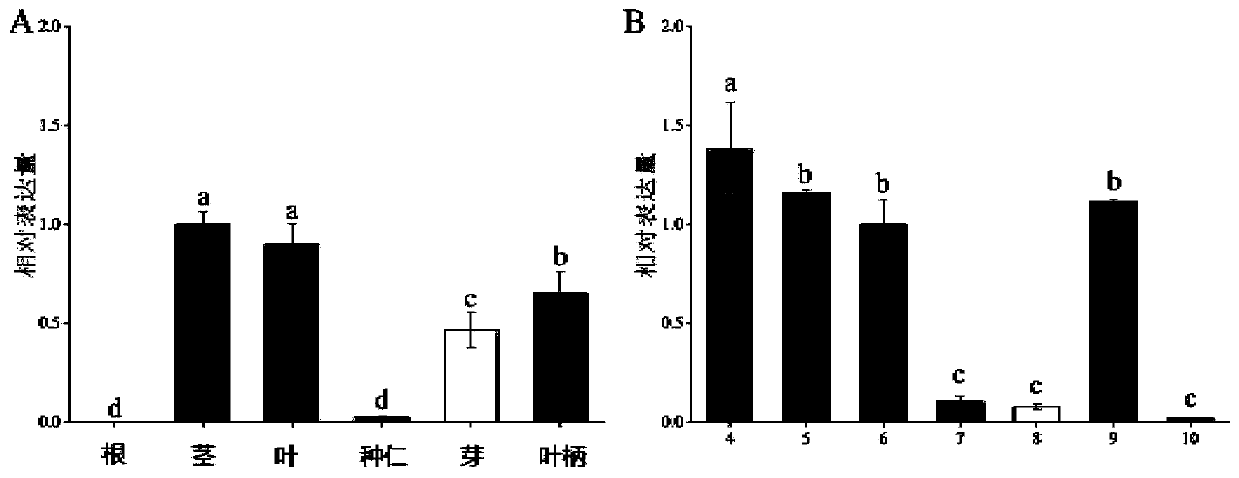
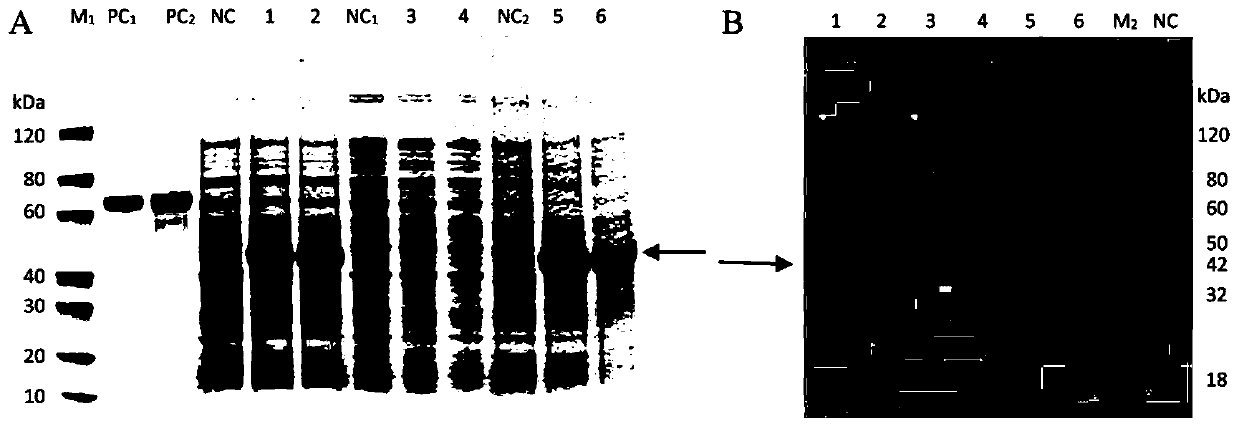
[0026] 1. Cloning of GbFLSa gene by RACE technology
[0027] Ginkgo DNA was extracted using the Plant Genomic DNA Kit (cetyltrimethylammonium bromide) (Zoman, Beijing, China). Total RNA was extracted from Ginkgo biloba leaves using RNAprep Pure Plant kit (Polysaccharides&Polyphenolics-rich, TIANGEN, Beijing, China). Based on Ginkgo biloba transcriptome data (NCBI Short Reads Archive database under access number SRP137637), specific primers for GbFLSa gene were designed, and the full-length cDNA sequence of GbFLSa was cloned by rapid amplification of cDNA ends (RACE). Nested primers were designed to amplify full-length cDNA using the SMATer RACE 5' / 3' Kit (Clontech, Palo Alto, CA, USA) kit. All primers were designed using Oligo 6.0 software (Table 1). It was amplified by PCR, recovered and purified by gel cutting, and transformed into E. coli competent cells through pMD19-T vector. Colonies were detected by PCR, and positive colonies were selected for Sanger sequencing. The...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com