High-sensitivity single-molecule RNA virus detection method based on RNA fluorescence in situ hybridization
A fluorescence in situ hybridization, RNA virus technology, applied in the detection of water and soil), clinical and environmental samples (such as food field, can solve the problem of unsatisfactory detection of virus accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0182] Embodiment 1: HEK293T cell culture and plasmid transfection test;
[0183] 12mm diameter coverslips were soaked and cleaned with 1N nitric acid as described above, and then coated with 0.01% polylysine (PDL; Sigma). HEK293T cells were seeded on coverslips and placed in 24-well cell culture plates at 37°C / 5% CO 2 Sterile constant temperature culture. The medium was MEM medium (GIBCO) plus 10% fetal bovine serum (FBS) and antibiotics (penicillin and streptomycin, GIBCO).
[0184] The plasmid uses SARS-CoV-2 (2019-nCoV) Nucleoprotein Gene ORF cDNA cloneexpression plasmid, C-OFPSpark tag (Codon Optimized; Sino Biological); wherein OFPSpark is a red (orange) fluorescent protein derived from DsRed (maximum excitation / emission wavelength 549 and 566nm, respectively). The protein is expressed in cells as a sign of successful transfection.
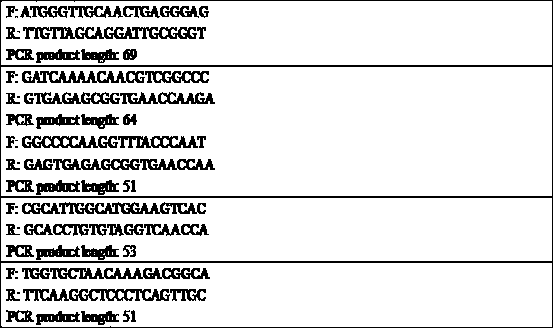
[0185] First, use Primer-Blast to design PCR primers for the nucleoprotein fragment of the plasmid, and set the length of the PCR produ...
Embodiment 2
[0195] Example 2: SARS-CoV-2 nasopharyngeal / oropharyngeal swab detection one
[0196] Use a special virus sampling tube to collect nasopharyngeal swabs / oropharyngeal swabs according to clinical requirements, make smears, and fix them with 4% PFA for 10 minutes. You can also shake the swab to elute the cells in the buffer, then use Thin Prep to make a smear, and fix the smear with 4% PFA for 10 minutes. The cells were then washed 2×5 minutes with PBS, followed by permeabilization with 0.2% Triton X-100 for 5 minutes, and then washed 1×5 minutes with PBS.
[0197] For each coverslip (or 1 cm on a microscope slide 2 smear area, the same below), use 40 μl of pre-hybridization solution or hybridization solution.
[0198] For each coverslip, prepare 20 μl of 80% deionized formamide / 1×SSC containing 1 μg salmon sperm ssDNA and 1 μg yeast tRNA, respectively. Heat this solution in a dry bath at 85°C for 5 min and place on ice. Then add 20μl hybridization buffer to get 40μl prehybri...
Embodiment 3
[0208] Example 3: SARS-CoV-2 Nasopharyngeal / Oropharyngeal Swab Detection II
[0209] Sample collection, processing and hybridization are the same as in Example 2.
[0210] Treat with PBS containing 0.2% hydrogen peroxide for 20 minutes, wash with PBS for 2×5 minutes, and perform the following steps.
[0211] Smears were blocked with 2% goat serum or 1% BSA for 20 minutes. Use polyclonal rabbit anti-DIG IgG antibody as primary antibody, incubate for one hour at room temperature, wash with PBS for 3×5 minutes, add secondary antibody goat anti-rabbit IgG-HRP, incubate for 30 minutes at room temperature, wash with PBS for 3×5 minutes, wash with AEC substrate Object color. After washing with PBS for 2×5 minutes, quickly stain with hematoxylin for nuclei, and then mount with water-soluble mounting solution (such as permanent immunohistochemical mounting medium VectaMount™). It can be inspected with ordinary light microscopes, and images can also be captured and analyzed with Leic...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com