Method for increasing yield of 2'-fucosyllactose in recombinant escherichia coli
A technology of fucosyllactose and Escherichia coli, which is applied in the field of genetic engineering, can solve the problems of high-efficiency expression transformation of FutC, unstable protein expression, inactive inclusion bodies, etc., and achieve the goal of simple construction method, promotion of synthesis, and improvement of transformation efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
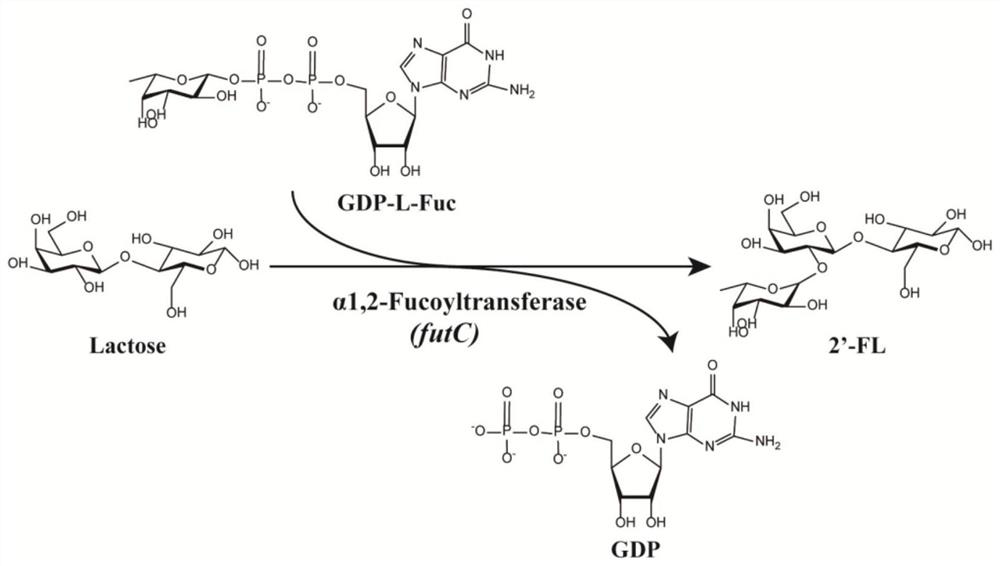
[0031] (1) Heterologous expression of α-1,2-fucosyltransferase derived from Helicobacter pylori
[0032] The futC sequence of the α-1,2-fucosyltransferase gene of Helicobacter pylori (ATCC No. 26695) published on NCBI was fully synthesized, and the futC gene and the expression vector pMA09 were amplified by PCR, using The template plasmid was digested with DpnI enzyme, and the futC gene fragment and the linearized vector pMA09 fragment were purified and recovered. The futC gene and the linearized vector pMA09 were then recombinantly ligated using a Seamless Cloning Kit (Seamless Cloning Kit). The recombination reaction system was reacted at 50°C for 15-20min, then ice-bathed for 30-40min, then the recombinant system was transferred to E. coli JM109 competent cells, recovered at 37°C for 1h, and coated on ampicillin-resistant LB with a final concentration of 0.1mM. Plates and incubated at 37°C for 10-12h. Finally, single colonies on the ampicillin-resistant plate were selecte...
Embodiment 2
[0045] Synthesis of 2'-FL by Shake Flask Fermentation Recombinant Bacteria
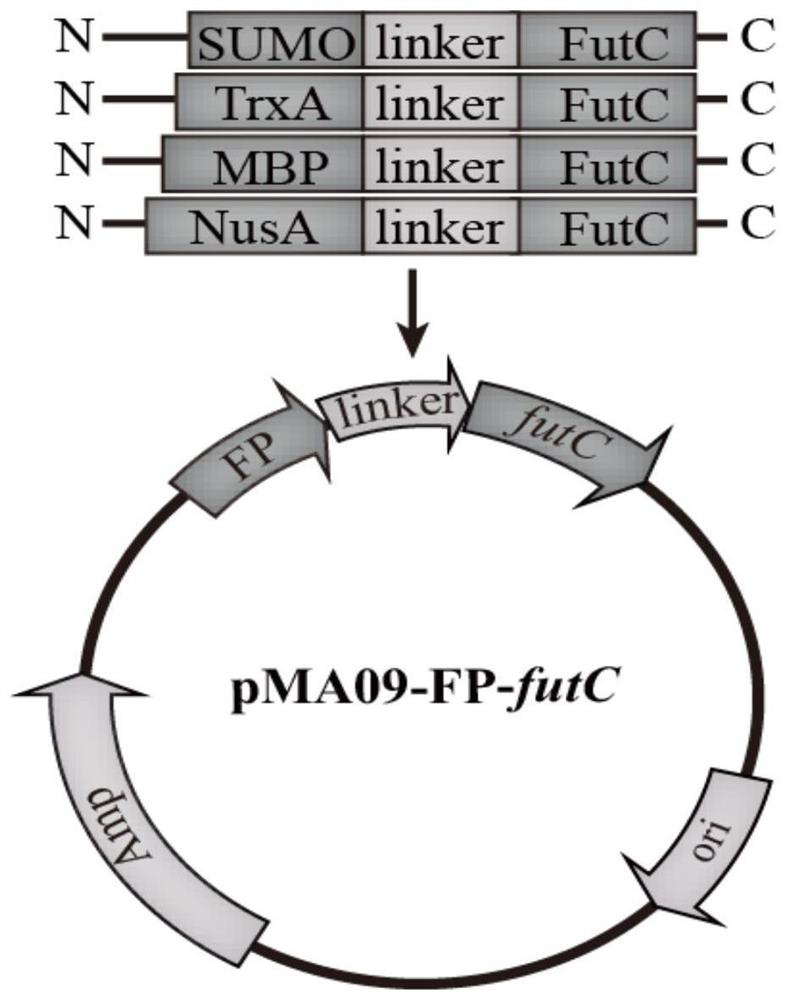
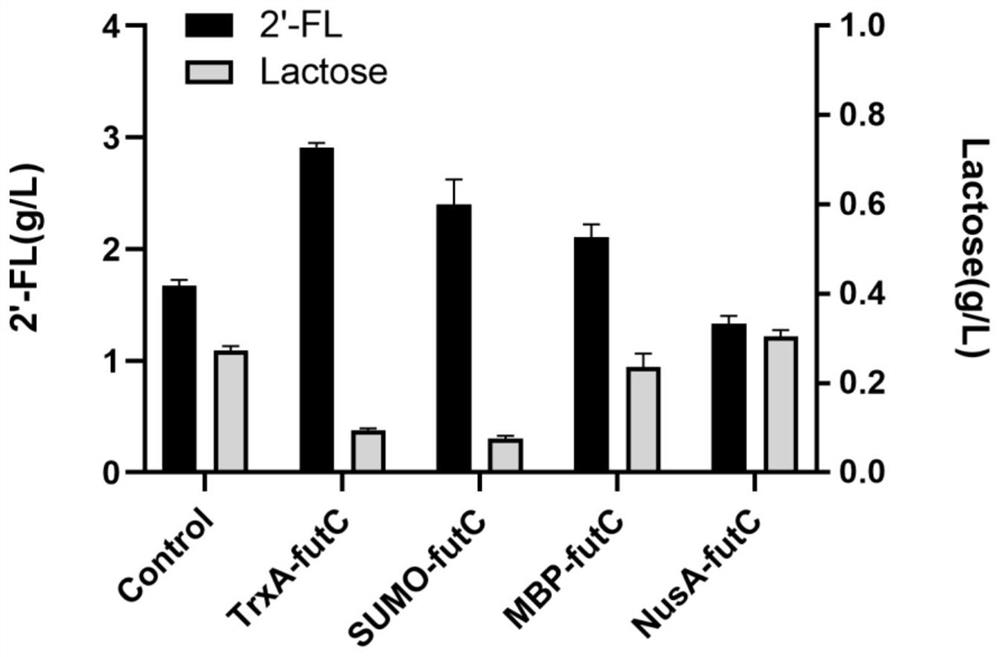
[0046]The recombinant plasmids pMA09-TrxA-futC, pMA09-SUMO-futC, pMA09-MBP-futC and pMA09-NusA-futC that were sequenced correctly in the above examples and comparative examples were transferred into MG-26 competent cells, and recovered at 37°C for 1 h. The ampicillin-resistant LB plate with a final concentration of 0.1 mM was cultured at 37°C for 10-12 h to obtain pMA09-FP-futC fermented recombinant bacteria fused with different protein tags. Pick a single colony into LB medium (tryptone 10g / L, yeast powder 5g / L, NaCl 10g / L) with a final concentration of 0.1mM ampicillin for 8-10h, and use it as the seed solution for shake flask fermentation. Then the seed liquid was inserted into a 250mL conical flask containing 20-25mL fermentation medium at an inoculum amount of 1%, and ampicillin with a final concentration of 0.1mM was added at the same time. The formula of the fermentation medium was: glycerol 5-...
Embodiment 3
[0049] Knock-in TrxA-futC fusion protein gene
[0050] (1) Preparation of E. coli MG-26 competent with pCas9 plasmid
[0051] First, the pCas9 plasmid was transformed into Escherichia coli MG-26 and spread on a 50 μg / mL kanamycin plate. After culturing at 30°C for 12 h, a single colony on the plate was picked, inoculated into fresh LB medium, and the final concentration was added. 50 μg / mL kanamycin antibiotic, cultured overnight at 30°C and 220 r / min; transfer the overnight cultured bacterial solution to a 250 mL conical flask containing 50 mL of LB medium at an inoculum of 1%, and wait for the bacterial cells OD 600 When it reaches 0.2, add arabinose with a final concentration of 30-40mmol / L to induce pCas9 plasmid to express recombinase; continue to culture to OD 600 When the temperature is 0.6-0.7, the bacterial solution was ice-bathed for 20 min, and the bacterial cells were collected by centrifugation at 5000 r / min; the cells were washed twice with pre-cooled sterilize...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com