Genetically engineered bacterium for producing N-acetylglucosamine and application of genetically engineered bacterium
A technology of genetically engineered bacteria and acetylamino, applied in the field of genetic engineering, can solve problems such as limiting the conversion rate of N-acetylglucosamine, and achieve the effects of strengthening industrial production potential, improving conversion rate, and improving synthesis efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
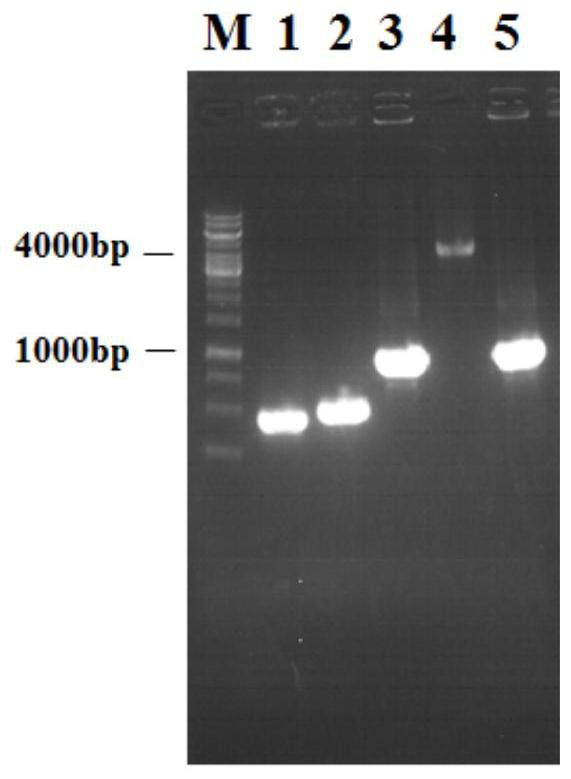
[0047] Embodiment 1 produces the construction of N-acetylglucosamine bacterial strain:
[0048] (1) Integration of T7 RNA polymerase gene (T7-RNAP)
[0049] The integration of T7 RNA polymerase gene (T7-RNAP) was carried out by Red recombination technology.
[0050] ①Using PCR technology to use the E.coli W3110 genome as a template, according to the xylose promoter P xylF Gene sequence design a pair of primers (PxylF-F / R) to amplify xylose promoter P xylF ;
[0051] ②Using PCR technology to use the E.coli BL21 genome as a template, design a pair of primers (T7-F / R) according to the T7 RNA polymerase gene sequence, and amplify the T7 RNA polymerase gene;
[0052] ③Using PCR technology to use the pKD3 plasmid as a template, design primers (Cm r -F / R) amplifying the chloramphenicol resistance gene fragment;
[0053] ④Using PCR technology to use the E.coli W3110 genome as a template, according to the LacIZ gene sequence, design upstream homology arm primers (T7-UF / UR) and dow...
Embodiment 2
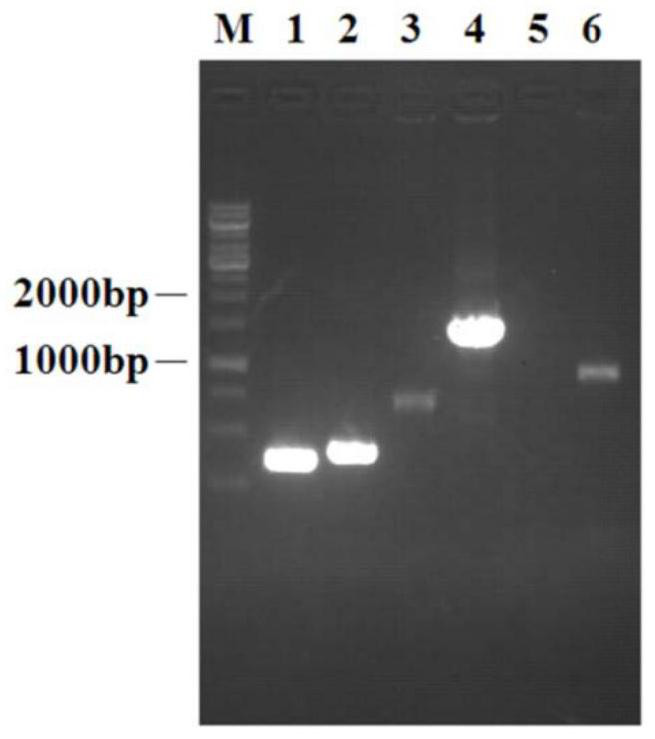
[0073] Example 2 Construction of N-acetylglucosamine-producing strains by complex carbon source metabolism division of labor
[0074] (1) Using CRISPR / Cas9 gene editing technology to knock out the pfkA gene
[0075] ①Using PCR technology to use the E.coli W3110 genome as a template, according to the pfkA gene sequence, design upstream homology arm primers (pfkA-UF / UR) and downstream homology arm primers (pfkA-DF / DR) at both ends of the gene, PCR Amplify and obtain the upstream and downstream homology arms of the pfkA gene;
[0076] ②Using overlapping PCR technology to use the upstream and downstream homology arms of the pfkA gene as templates, PCR amplification was used to obtain overlapping fragments of the upstream and downstream homology arms of the pfkA gene;
[0077]③ Construct a gRNA plasmid containing the Cas9 cleavage recognition sequence. The DNA fragment containing the target sequence is obtained by annealing the primers pfkA-F' and pfkA-R'. The constructed gRNA pla...
Embodiment 3
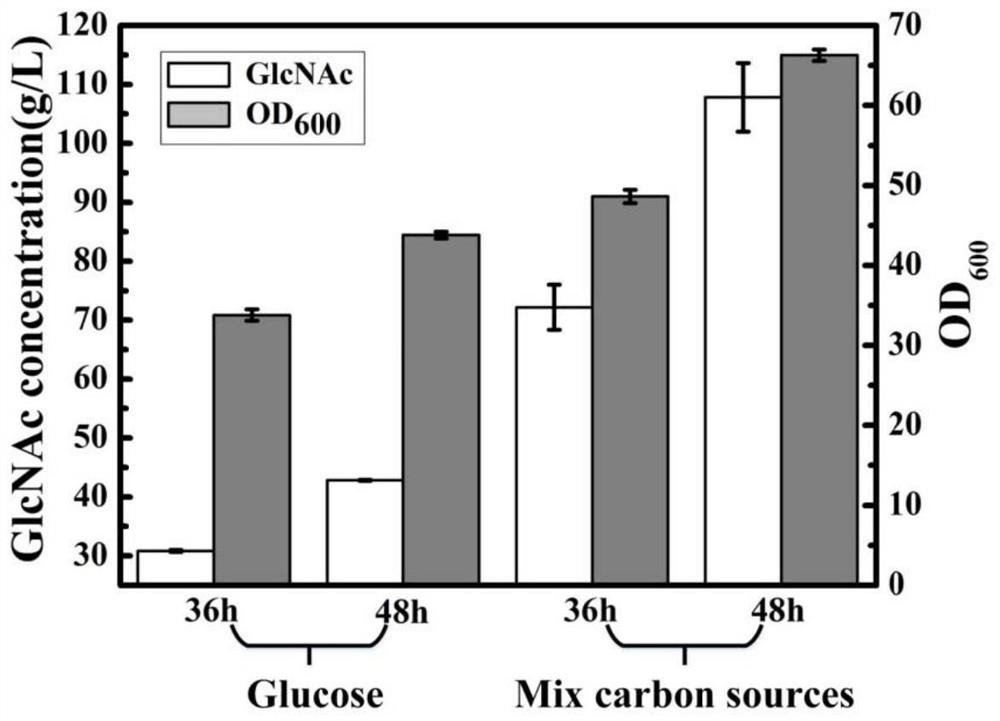
[0089] Embodiment 3 shaking flask fermentation experiment
[0090] Produce N-acetylglucosamine with the bacterial strain constructed in the above-mentioned embodiment 2 as a production strain:
[0091] (1) Seed culture: Use an inoculation loop to scrape a ring of strains into a 500 mL shaker flask with a liquid volume of 30 mL, and culture at 37° C. and 220 rpm for 12 hours.
[0092] (2) Shake flask fermentation: inoculate the fermentation medium with 10% inoculation amount, cultivate for 48 hours at 37° C. at 220 rpm; maintain the pH at 7.0-7.2, and maintain the fermentation with 60% (m / v) glycerol glucose composite carbon source (using phenol red As an indicator, if the color of the fermentation broth remains unchanged, it is considered to be short of sugar. During the lack of sugar, 60% (m / v) glycerol glucose composite carbon source) is added, and a xylose solution with a final concentration of 10g / L is added to the initial fermentation to induce the expression of the targe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com