Engineering probiotic with surface display phenylalanine ammonia lyase
A phenylalanine ammonia lyase, surface display technology, applied in the field of genetic engineering, can solve the problems of long-term persistence, low transport efficiency, heavy economic burden, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
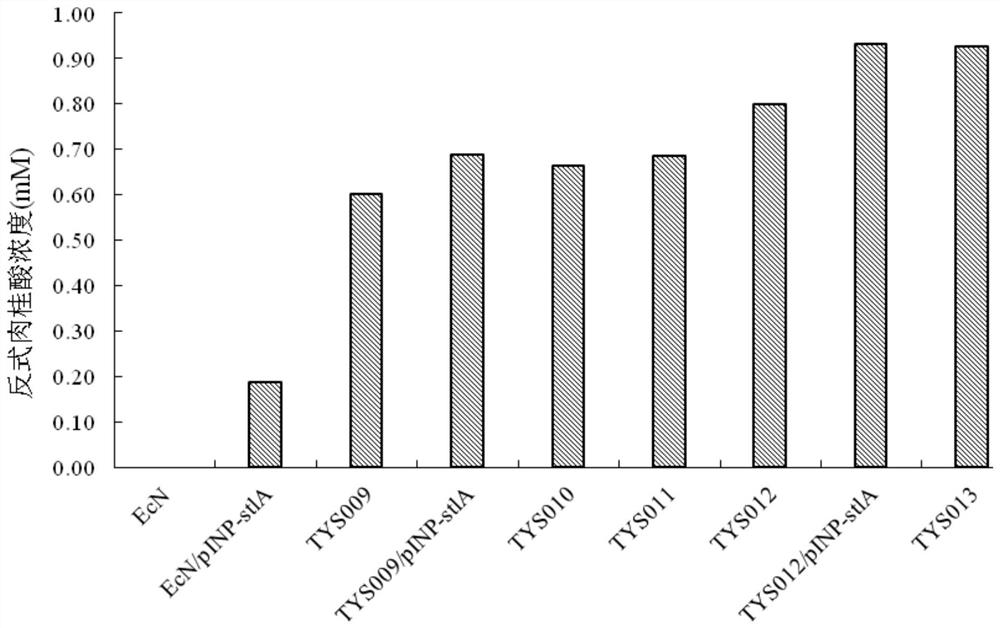
[0066] Embodiment 1: Construction of surface display PAL engineering probiotics EcN / pINP-stlA
[0067] 1.1 pINP-stlA plasmid construction
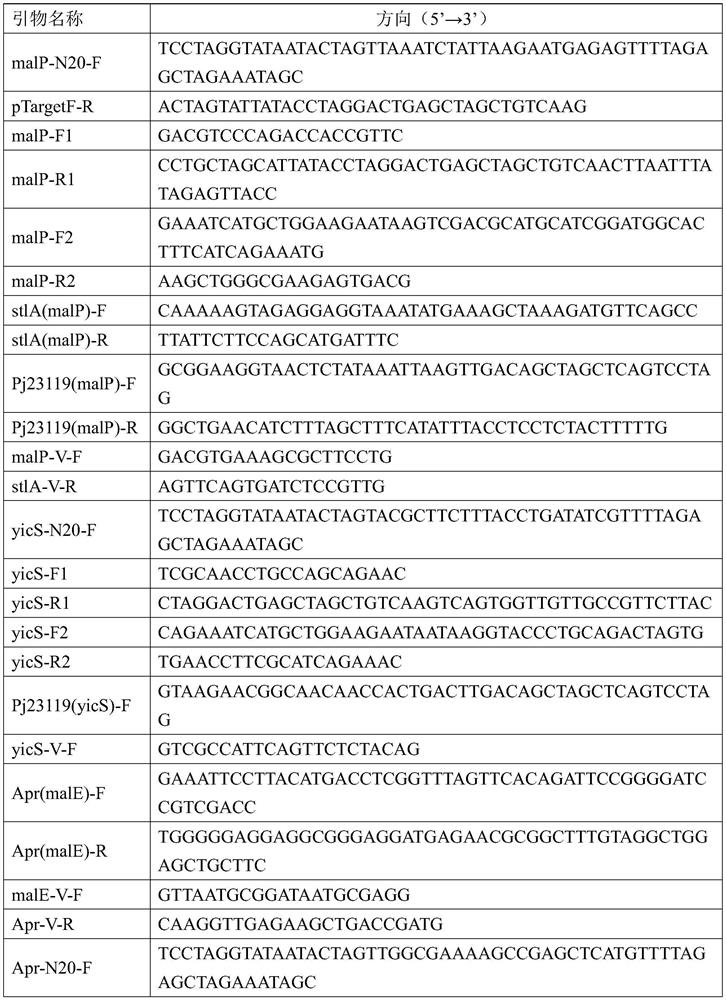
[0068] (1) Using the pSU2718 plasmid as a template and 15A-F / Psu-RG as primers, PCR amplifies the 15A fragment, about 1kb; using the pPIC9k plasmid as a template, Kan-FG / Kan-R(15A) as primers, PCR amplification Kan fragment, about 1kb; with pUC-inak plasmid as template (synthesized by GenScript), inak-F / inaK-R as primers, inak-N fragment was obtained by PCR amplification, about 600bp; pUC-sltA plasmid (GenScript Rui Synthetic) as template, stlA(inaK)-F / stlA-R as primers, PCR amplified stlA(inaK) fragment, about 1.6kb; using pTrc99a plasmid as template, rrnB-F / rrnB-R as primers, PCR amplified The rrnB fragment was obtained by increasing, about 400bp.
[0069] (2) 15A, Kan, inak-N, stlA (inaK) and rrnB fragments were ligated using the DNA assembly method (DNA assembly Kit was purchased from Quanshijin), and the ligated products were transf...
Embodiment 2
[0073] Example 2: Construction of engineering probiotics TYS009 and TYS009 / pINP-stlA for degrading phenylalanine
[0074] 2.1 pTargetF-malP plasmid construction
[0075] Using the pTargetF plasmid (Addgene: 62226) as a template and malP-N20-F / pTargetF-R as primers, PCR amplifies the malP-N20 fragment, about 2.2kb, digests the PCR fragment with DpnI, and transforms it into Escherichia coli DH5α chemical sensory The pTargetF-malP plasmid was obtained by screening on LB solid plates containing spectinomycin (50 μg / mL) at 37°C in competent cells; for the preparation method of chemical transformation competent cells, refer to "Molecular Cloning Experiment Guide" (third edition ).
[0076] 2.2 malP site knock-in Pj23119-stlA
[0077] (1) Electroporation fragment preparation: E.coli Nissle 1917 genome was used as template, malP-F1 / malP-R1, malP-F2 / malP-R2 were used as primers, and malP-UP and malP-DN fragments were obtained by PCR amplification respectively , about 600bp respectivel...
Embodiment 3
[0140] Example 3: Construction of arginine pathway enhanced engineering probiotic TYS010
[0141] 3.1 Knock-in of apramycin resistance gene at argA site
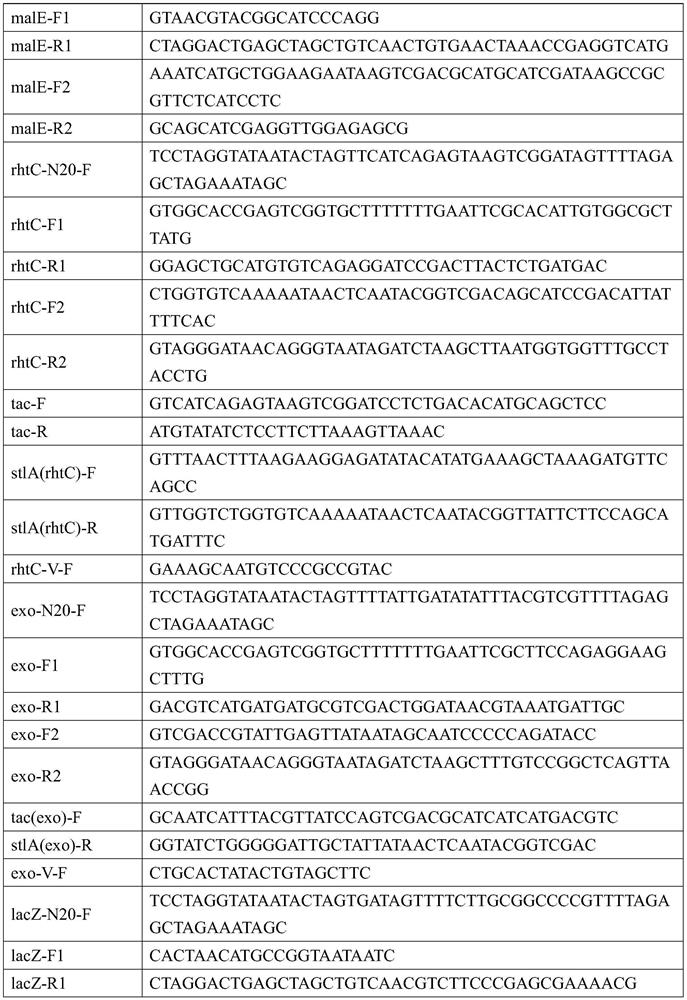
[0142] (1) Amplification of fragments containing homology arms: pIJ773 plasmid (Gust B, et al., PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin.Proc.Natl.Acad.Sci.U.S.A.2003 , 100:1541-1546.) as a template, Apr(argA)-F / Apr(argA)-R as primers, PCR amplifies the Apr(argA) fragment, about 1.4kb, and DpnI digests the PCR fragment;
[0143] (2) Electroporation: The method is the same as 2.2, and the Apr(argA) fragment is electrotransformed into EcN (malP::Pj23119-stlA, yicS::Pj23119-stlA, malE::Pj23119-stlA, rthC::Ptac-stlA obtained in Example 2 , exo::Ptac-stlA, lacZ::Pj23119-pheP, agaI::Pj23119-pheP, araBD::Para-pma,△dapA) / pCas competent cells, coated with apramycin (50μg / ml), kanamycin (50 μg / ml) and diaminopimelic acid (100 μg / mL), cultiv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com