Mutant enzyme CYP153A M228L and application thereof in synthesis of 10-hydroxy-2-decenoic acid
A technology of mutating enzymes and hydroxyl groups, applied in the field of biological fermentation, can solve the problems of not reaching the industrialized level, low synthesis efficiency, etc., and achieve the effect of improving the conversion rate and high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
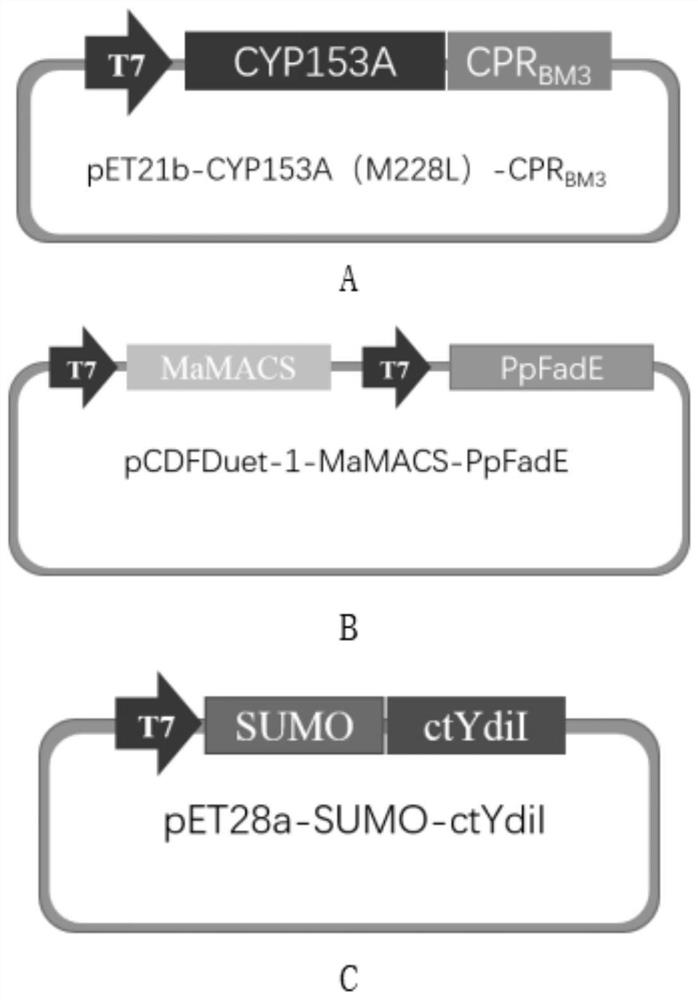
[0104] The fusion enzyme gene of alkane hydroxylase CYP153A of Marinobacter aquaeolei and P450NADH reductase of Bacillus megaterium, namely CYP153A-CPR BM3 Fusion enzyme gene, CYP153A M228L-CPR BM3 PCR amplification of fusion enzyme gene, acyl-CoA dehydrogenase gene PpFadE, acyl-CoA synthetase gene MaMACS, and acyl-CoA thioesterase gene ctydiI.
[0105] According to the fusion enzyme gene of alkane hydroxylase CYP153A of Marinobacter aquaeolei and P450NADH reductase of Bacillus megaterium, that is, CYP153A gene, CPR BM3 Gene, CYP153A M228L-CPR BM3 Fusion enzyme gene, acyl-CoA dehydrogenase gene PpFadE, acyl-CoA synthetase gene MaMACS, acyl-CoA thioesterase gene ctydiI design PCR amplification primers, the nucleotide sequences of the upstream primers are respectively as SEQID NO.1, 3, 5, 7, 9, and 11, the nucleotide sequences of the downstream primers are shown in SEQ ID NO.2, 4, 6, 8, 10, and 12;
[0106] Among them, the fusion enzyme gene of alkane hydroxylase CYP153A of M...
Embodiment 2
[0155] Digestion of recombinant plasmid vectors pET21b, pCDFDuet-1, pCDFDuet-1-MaMACS, pET28a-SUMO and gene ctYdiI
[0156] Extract the pET21b plasmid and perform a double enzyme digestion reaction. The reaction system is as follows:
[0157]
[0158] Reaction conditions: react at 37°C for 6h.
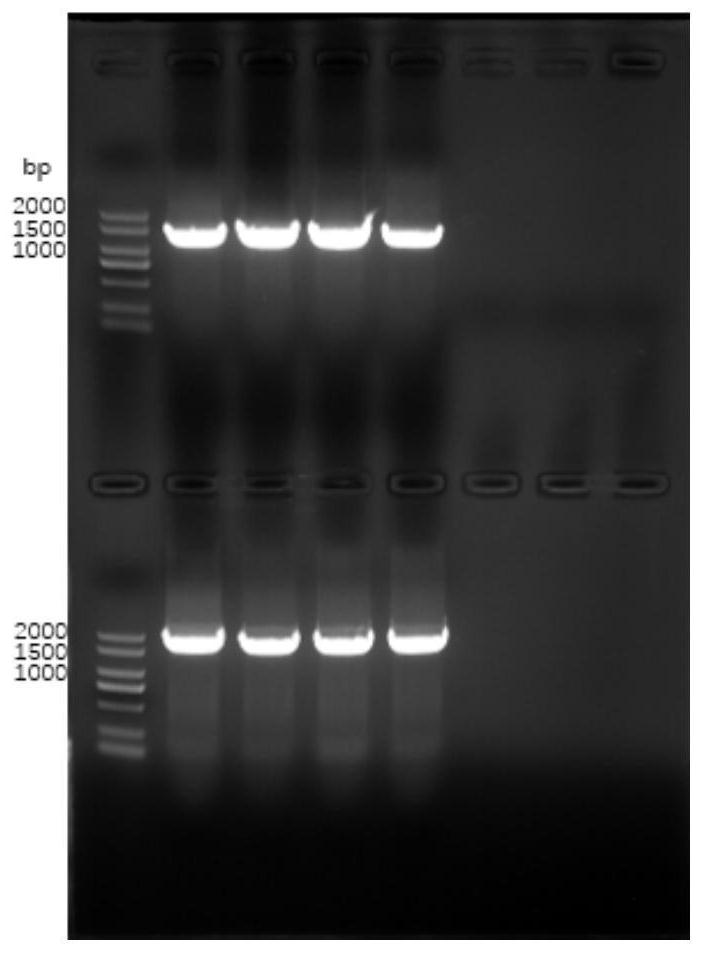
[0159] The pET21b plasmid vector was purified by 1% agarose gel electrophoresis after double digestion Figure 5 , and use the DNA gel recovery kit to recover the target fragment.
[0160] Extract the pCDFDuet-1 plasmid and perform a double enzyme digestion reaction, the reaction system is as follows:
[0161]
[0162] Reaction conditions: react at 37°C for 6h.
[0163] The pCDFDuet-1 plasmid vector was double digested and purified by 1% agarose gel electrophoresis, and the target fragment was recovered using a DNA gel recovery kit.
[0164] Extract pCDFDuet-1-MaMACS plasmid and perform double enzyme digestion reaction on it, the reaction system is as follows:
[0165]
...
Embodiment 3
[0173] Recombinant plasmid pET21b-CYP153A-CPR BM3 Multi-fragment seamless cloning, recombinant plasmid pET28a-SUMO-ctYdiI connection
[0174] Combine the double digested pET21b plasmid with CYP153A, CPR BM3 The PCR product is ligated, and the ligation reaction system is as follows:
[0175]
[0176] Mix the above ligation reaction system thoroughly and centrifuge for a few seconds, collect the droplet on the tube wall to the bottom of the tube, and connect at 37°C for 30 minutes to obtain the recombinant plasmid pET21b-CYP153A-CPR BM3 .
[0177] Ligate the digested pCDFDuet-1 plasmid with the MaMACS PCR product, and the ligation reaction system is as follows:
[0178]
[0179]The above ligation reaction system was thoroughly mixed and then centrifuged for a few seconds to collect the drop on the tube wall to the bottom of the tube, and reacted at 37°C for 37 minutes to obtain the recombinant plasmid pCDFDuet-1-MaMACS.
[0180] Ligate the digested pCDFDuet-1-MaMACS p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com