Genetically engineered bacterium for selectively producing retinol as well as construction method and application thereof
A technology of genetically engineered bacteria and retinol, applied in the fields of metabolic engineering, microorganisms, and genetic engineering, can solve the problem of low selectivity of all-trans isomers, and achieve the effects of increasing yield, good application prospect and yield improvement.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1. Cloning of genes required for retinol biosynthesis
[0053] 1. Escherichia coli and Saccharomyces cerevisiae genomic DNA extraction
[0054] 1.1 The extraction of Escherichia coli BL21 genomic DNA is completed by the kit, and the specific steps are as follows:
[0055] (1) Take 1 mL of Escherichia coli cultured overnight, add it to a 1.5 mL centrifuge tube, centrifuge at room temperature at 8000 rpm for 1 min, discard the supernatant, and collect the cells. Add 180 μL of lysozyme solution to resuspend the bacteria, and bathe in water at 37°C for 30-60 minutes. Then add 20 μL Proteinase K solution, shake and mix. Water bath at 56°C for 30 min until the cells were completely lysed.
[0056] (2) During the water bath process, mix by inverting every 10 minutes until the mixture becomes clear and transparent, add 20 μL of RNase, and place at room temperature for 2-5 minutes to remove RNA.
[0057] (3) Add 200μL Buffer BD and mix thoroughly by inversion.
[00...
Embodiment 2
[0086] Example 2. Construction of pUMRI-MOT3 plasmid
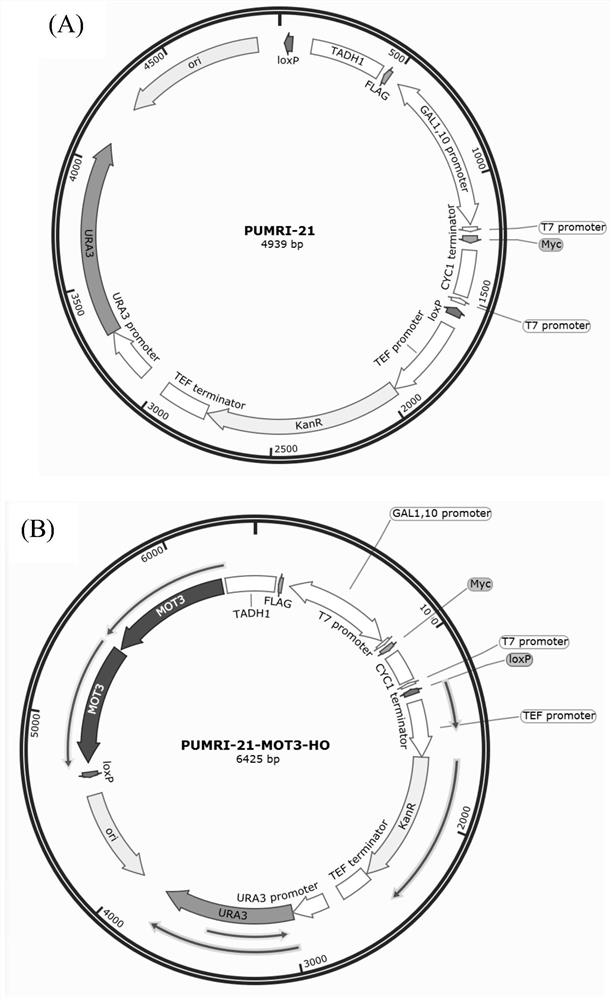
[0087] The upstream and downstream homology arms of the transcriptional repressor (MOT3) used to construct the pUMRI-MOT3 plasmid use the yeast genome as a template and the backbone part of the pUMRI-MOT3 plasmid uses the pUMRI21 plasmid as a template, using the high-fidelity enzyme PrimeSTARTM HS DNA polymerase) PCR amplification was carried out, and the three fragments were connected by Gibson Assembly kit. build like figure 1 shown. The nucleotide sequence of the mevalonate pathway transcription inhibitor gene is shown in SEQ ID NO.8.
[0088] The specific primer design is as follows:
[0089] Table 2 Primers used in pUMRI-MOT3 plasmid construction
[0090]
Embodiment 3
[0091] Example 3. Construction of plasmids required for the retinol biosynthetic pathway
[0092] 1. Enzyme digestion and gel recovery
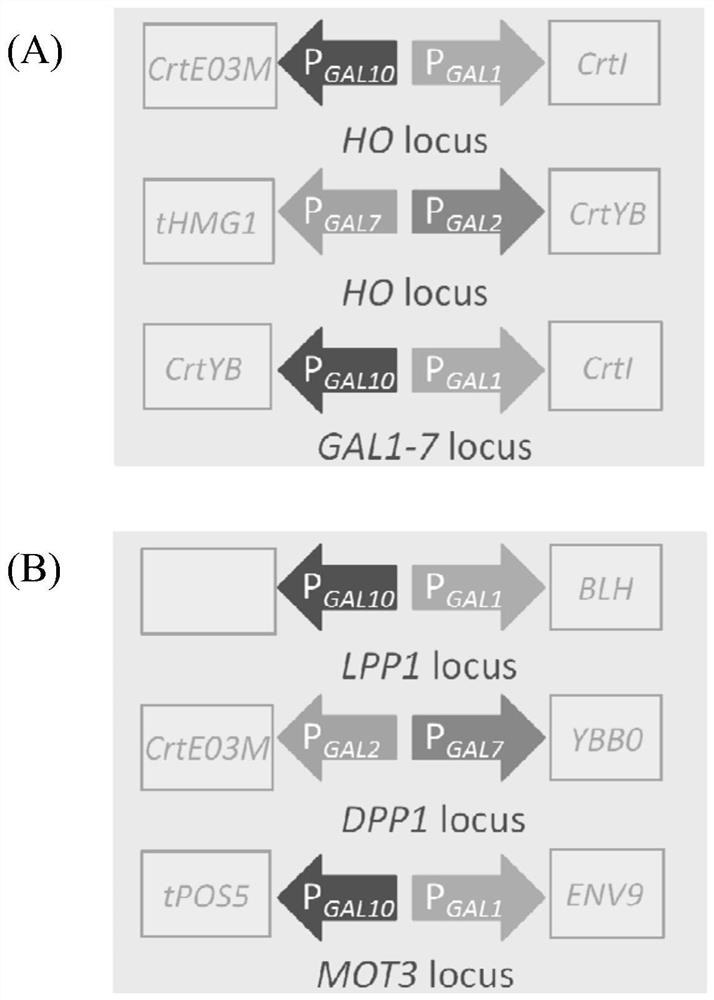
[0093] The pUMRI series integration plasmids (pUMRI-LPP1 and pUMRI-DPP1 were both constructed and preserved by the laboratory, see Example 2 for the construction of pUMRI-MOT3) and the target fragment of the PCR product were double-digested by Takara restriction endonuclease, double-digested The system followed the instructions of Takara restriction endonuclease, and after digestion, the system was subjected to DNA gel recovery treatment, and the specific steps were carried out according to the instructions of the Axygen kit.
[0094] 2. Enzyme linkage
[0095] The digested fragments and plasmids were ligated using T4 DNA ligase, and the ligation system (10 μl) was as follows:
[0096]
[0097]
[0098] 22°C, connect for 30min.
[0099] 3. Conversion
[0100] Add 10 μL of the ligation product to the E. coli competent solution, place...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com