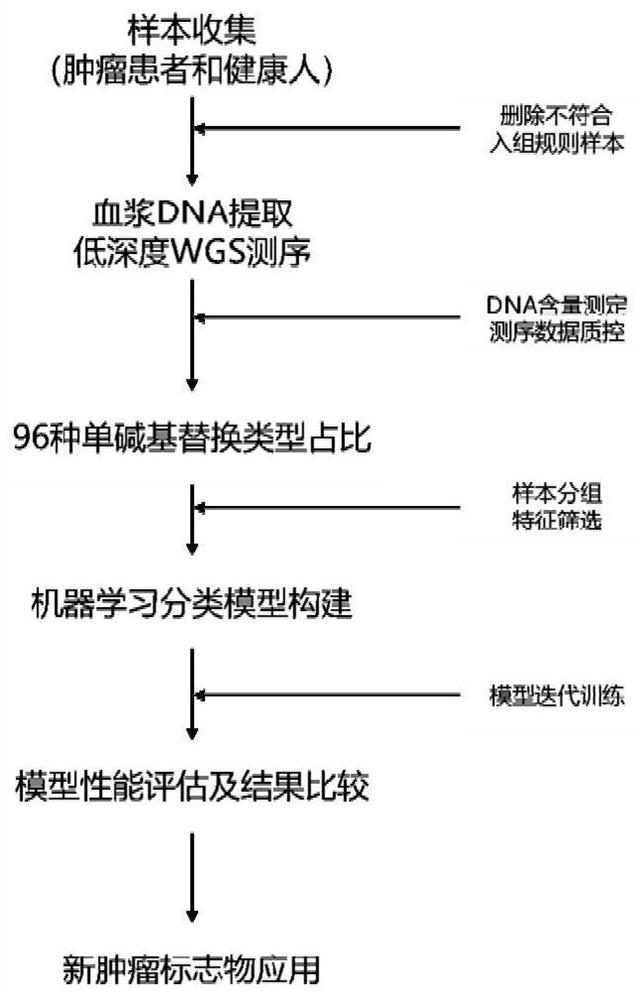
Tumor marker screening method based on single base substitution characteristic and application
A technology of tumor markers and screening methods, applied in the field of quantification of related single base substitution features, can solve problems such as inability to find, different sites and types, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] The enrolled samples of this embodiment come from another study of the inventor, in which several ideas of the inventor were verified simultaneously, and a total of 957 subjects were included, specifically 481 patients with liver cancer (HCC) and 476 patients. healthy control (NC) ( figure 1 ). Follow the steps below to extract plasma cell-free DNA (cfDNA):
[0080](1) Take 3ml of peripheral blood (collected and stored in Streck cell-free DNA blood collection tubes), Eppendorf centrifuge (5810R and 5427R, German), and centrifuge at 1600g at low speed for 10min at a low temperature of 4°C, and only take the supernatant; Then centrifuge at a high speed of 16000 g for 10 min, and take the supernatant to obtain the plasma sample. The cell-free DNA in the plasma was extracted with the kit MagMAXCell-Free DNA Isolation Kit (Thermo) and the nucleic acid extractor (Thermo Kingfisher FLEX, USA).
[0081] (2) DNA quality detection: DNA concentration was detected by Qubit 3 Nu...
Embodiment 2
[0083] Low-depth whole-genome sequencing was performed on the cfDNA samples of all subjects prepared in Example 1. The sequencing process is as follows:
[0084] (1) WGS library construction and on-machine sequencing: Take 5ng cfDNA to construct a pre-library with an Enzymatics (USA) related kit, mainly including two steps of end repair (5X ER / A-Tailing Enzyme Mix) and adapter (WGSLigase) , the adapter sequence is suitable for the IlluminaNovaSeq 6000 sequencing platform. The adapters were ligated and purified using XP magnetic beads (Agencourt AMPure XP beads, Beckman Coulter). The concentration value of WGS library was determined by qPCR (KAPALibrary Quant Kit, Roche), and the library size was determined by Fragment Analyzer (Agilent, USA). Afterwards, paired-end 150bp sequencing was performed on the IlluminaNovaSeq 6000 sequencing platform, and the average data volume of a single sample was 2X that of the whole genome.
[0085] (2) Data quality control: Use Fastp softwar...
Embodiment 3
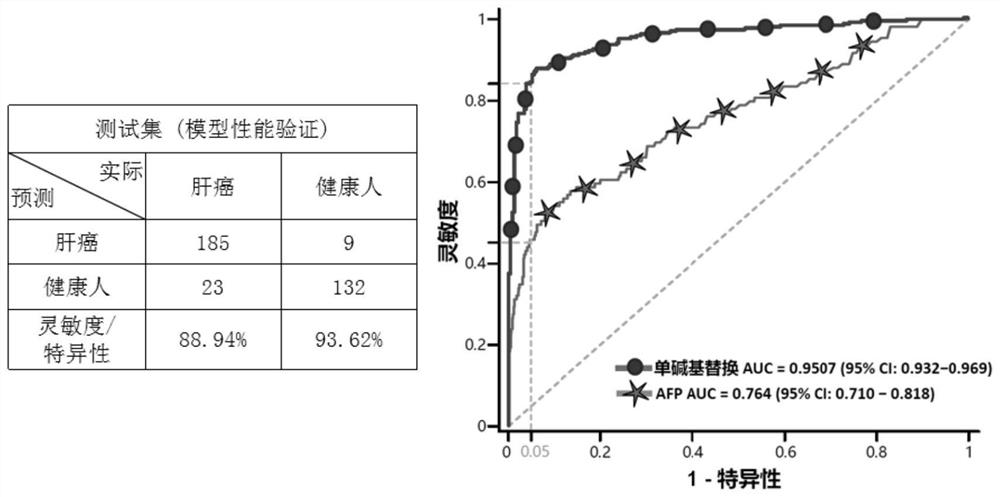
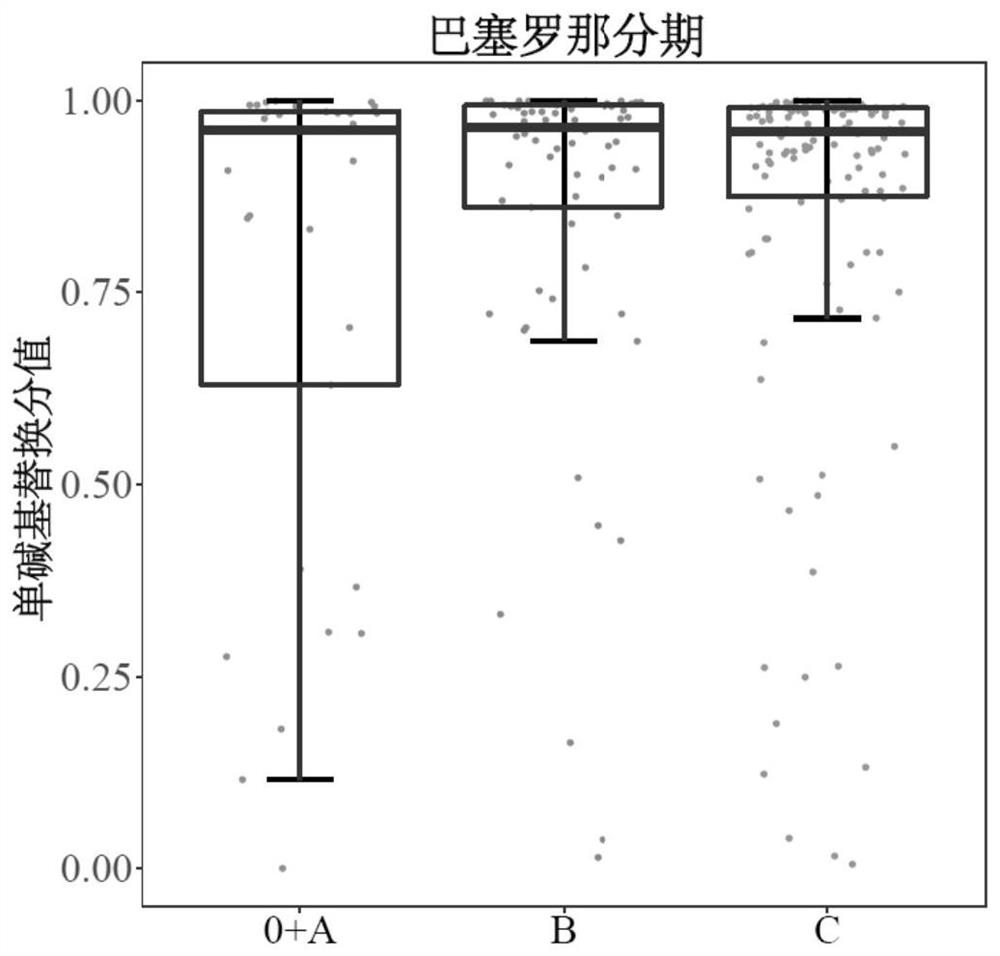
[0087] The sequencing data of tumor patients and healthy people were divided into independent training set (510 cases), validation set (98 cases) and test set (349 cases). The training set is used to screen the characteristic single base substitution type, the validation set is used to determine the optimal threshold of the model, and the test set is used to evaluate the model performance.
[0088] Based on the sequencing data obtained in Example 2, calculate the proportion of each type of single-base substitution in each training set sample: use Varscan software to output all sites with single-base substitution mutations in the whole genome, and select the mutation support number (that is, the same base substitution support number) is greater than or equal to 2 sites, remove the high-frequency mutation sites included in the dbSNP database, so as to reduce the impact of background mutations as much as possible, and reduce the systematic error to a certain extent. influences. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com