Method for diagnosing cancer using cfdna
A cancer cell, prostate cancer technology, applied in the field of detecting a biomarker gene specifically expressed or overexpressed in cancer by using the device, can solve the problem of spending a lot of time, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example 1
[0584] Preparation example 1. Preparation of nanowires whose surface is treated with cationic polymer
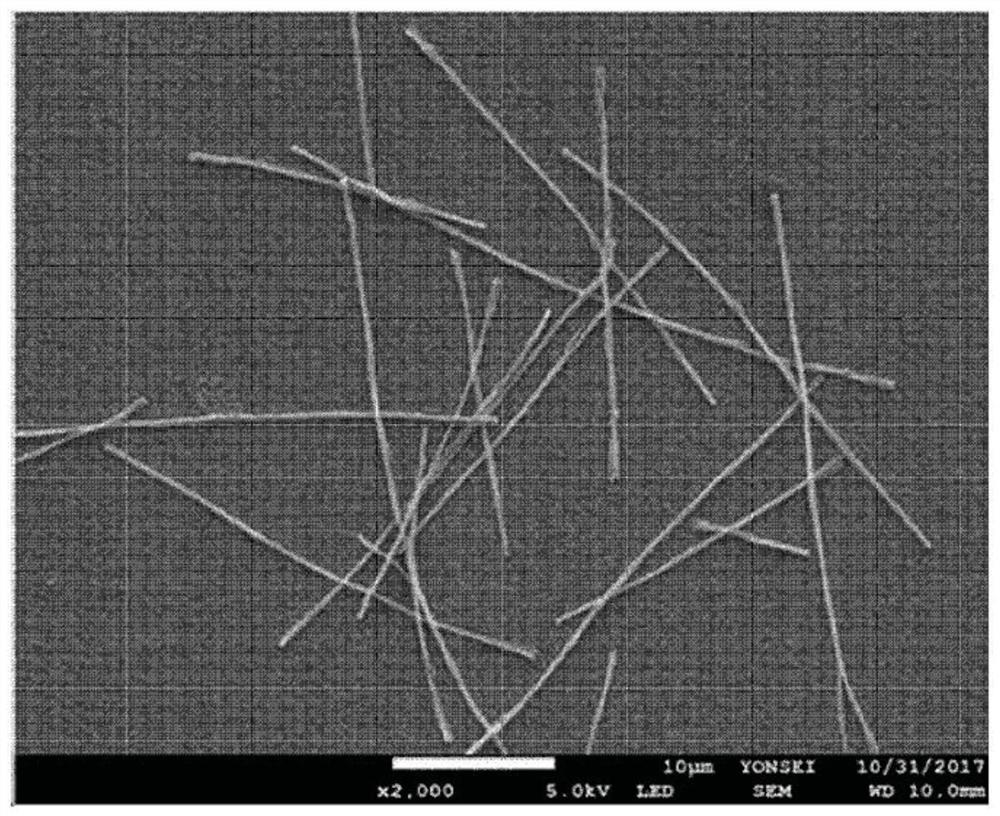
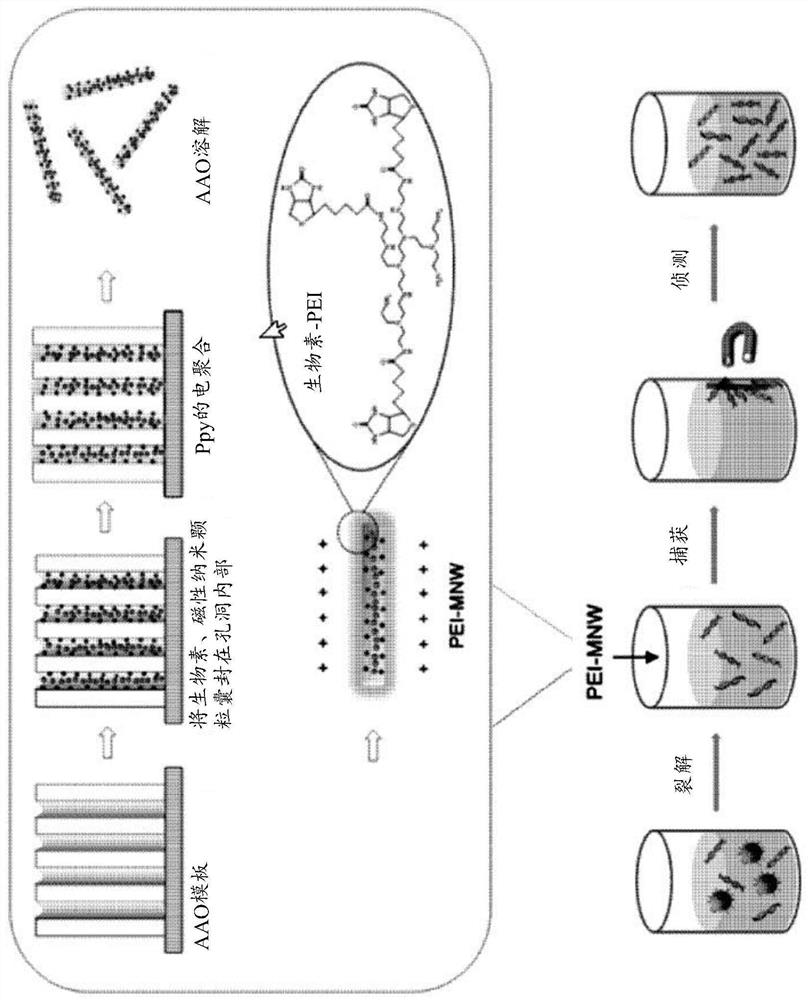
[0585] Such as Figure 1a As shown, nanowires with cationic polymer polyethyleneimine (PEI) conjugated to the nanowire surface were prepared. One surface of anodized aluminum oxide (AAO) was coated with a Q150T modular coating system (Quorum Technologies, UK) at 5 × 10 -3 A gold (Au) layer (thickness about 150 nm) was applied at mbar and 50 mA for 600 seconds. All electrochemical experiments were performed on AAO templates coated with gold (Au) using a potentiostat / galvanostat (BioLogic SP-150) equipped with a platinum wire counter electrode and an Ag / AgCl (3.0M NaCl type) reference electrode. .
[0586] To prepare cationic polymer-treated nanowires (PEI / Ppy NWs) together with 0.01 M poly(4-styrenesulfonic acid) and 0.01 M pyrrole solution containing 1 mg / ml biotin, chronoamperometry was performed at 1.0 V ( vs. Ag / AgCl) was applied to the pores of the AAO template for 7 ...
preparation example 2
[0589] Preparation example 2. Preparation of nanowires whose surface is treated with cationic polymer
[0590] According to substantially the same method as in Preparation Example 1 above, a nanostructure (PL / Ppy NW) having polylysine but not polyethyleneimine conjugated to the surface of the nanostructure was obtained.
preparation example 3
[0591] Preparation example 3. Preparation of poly(pyrrole) nanoparticles labeled with HRP and streptavidin
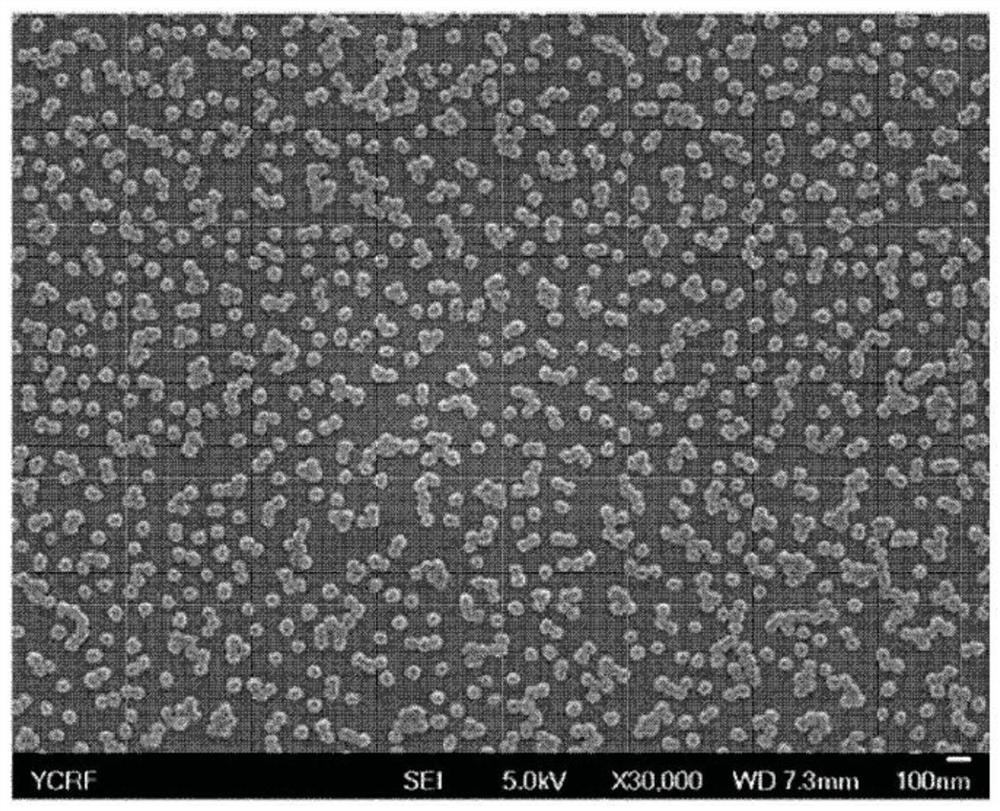
[0592] To prepare nanoparticles conjugated with HRP and streptavidin, 0.5 g of polyvinylpyrrolidone (PVP) was added to 12.5 mL of tertiary distilled water, stirred for 30 minutes, then 65 μL of pyrrole was added, and stirred for another 10 minutes. Afterwards, add 0.5 mL of FeCl at a concentration of 0.75 g / mL 3 solution and reacted for 3 hours. Afterwards, 20 mL of hyaluronic acid aqueous solution (400 mg / 20 mL) was added and stirred for 3 hours to prepare poly(pyrrole)-hyaluronic acid nanoparticles (Ppy-HA-NPs).
[0593] MWCO: Dialysis was performed for two days in triple distilled water using a 50,000 pore size membrane. Centrifuge at 1,200 rpm for 3 minutes to remove large particle aggregates, then lyophilize. Add 200 μg of Ppy-HA-NPs prepared above to 1 mL of triple distilled water, then add 100 mM EDC / 50 mM NHS solution and react for 45 minutes to activate the ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com