Gibberella fujikuroi genetically engineered bacterium with high-yield gibberellin GA3, construction method and application
A technology of genetically engineered bacteria, Gibberella fujikura, applied in the field of metabolic engineering, can solve the problems such as the inability of the strain to synthesize uracil and the inability of the initial strain to survive, and achieve the effects of releasing transcription inhibition, weakening feedback inhibition and efficient fermentation production.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Acquisition of F. fujikuroi-ΔpyrG
[0032] Knockout of the pyrG gene:
[0033] In order to reduce the interference of negative transformants and improve the selection efficiency of transformation, the pyrG gene of Gibberella Fujikura was knocked out, see Van Hartingsveldt W, Mattern I E, van Zeijl C M J, etal.Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Molecular and General Genetics MGG,1987,206(1):71-75.
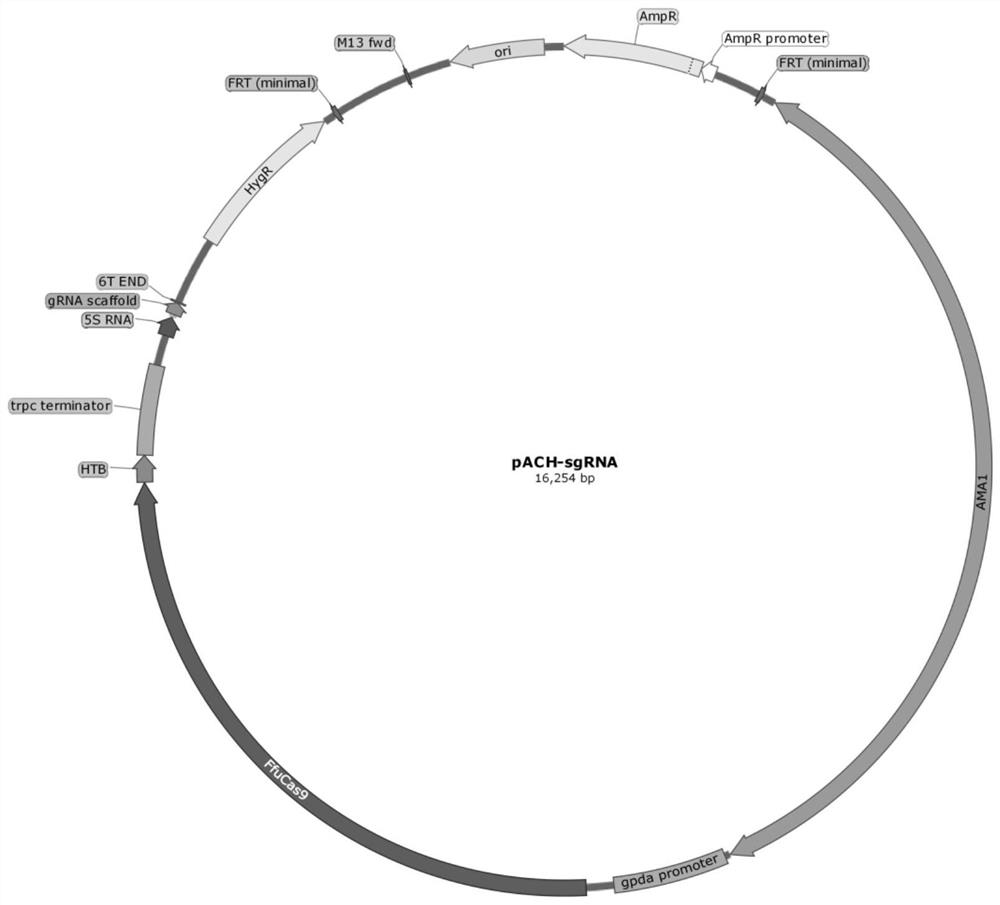
[0034] The pyrG gene encoding orotidine-5'-phosphate decarboxylase in the genome of Gibberella fujikuroi (Fusarium fujikuroi) CCTCC NO: M2019378 (disclosed in CN110527630A) was knocked out by CRISPR-Cas9 system. Using primer 1 and primer 2 by PCR, the sgRNA-ΔpyrG-1 fragment was amplified using the sgRNA expression cassette as a template. The PCR reaction conditions were as follows: 98°C for 3min; 98°C for 15s, 60°C for 15s, and 72°C for 15s, repeating 30 cycles ; Continue at 72°C for 3 minutes.
[0035...
Embodiment 2
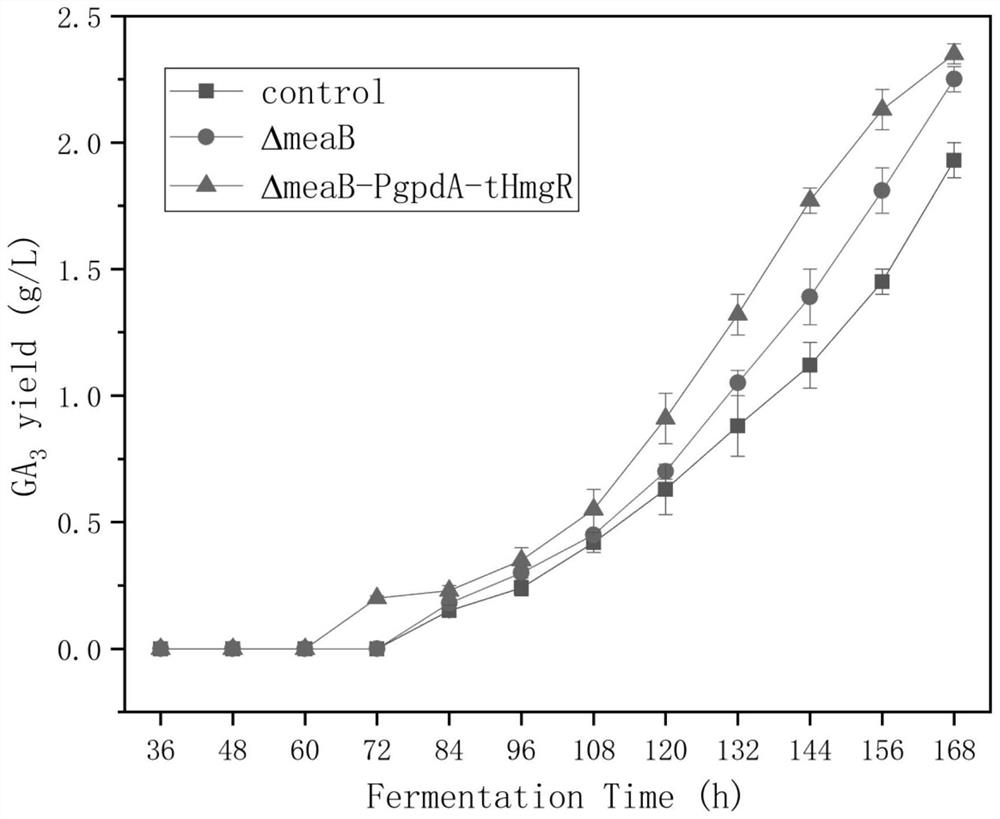
[0041] Example 2 F. Acquisition of Fujikuroi-RE:pyrGΔmeaB
[0042] (1) Completion of pyrG gene
[0043] The pyrG-deficient Gibberella fujikura cannot survive due to the inability to synthesize uracil. By complementing the pyrG gene, Gibberella can regain the ability to synthesize uracil, resulting in a reversion mutant strain that can survive on a plate without exogenous uracil . Using primers 11 and 12 by PCR, the complete pyrG expression cassette was amplified using the genome of Fusarium fujikuroi CCTCC NO: M2019378 as a template. The PCR reaction conditions were as follows: 98°C for 5min; 98°C for 30s, 60°C for 30s, 72°C for 1min30s, repeat 30 cycles; continue at 72°C for 5min. The PCR products were detected by 1.0% agarose gel electrophoresis and the purified fragments were recovered by cutting the gel.
[0044] (2) Amplification of upstream and downstream homology arms of meaB gene
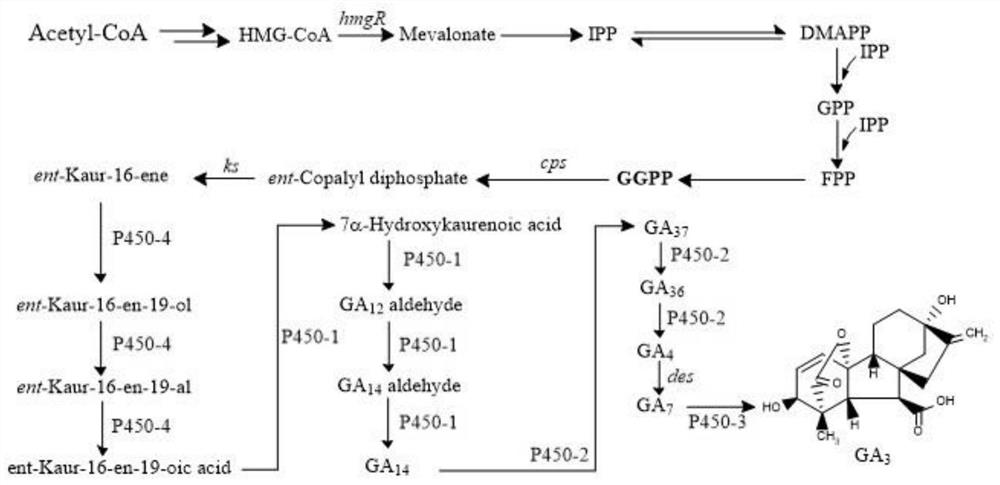
[0045] The bZIP-type transcriptional repressor meaB represses the expression of the ...
Embodiment 3
[0053] Example 3: Acquisition of F.fujikuroi-RE:pyrG GpdA-tHmgRΔmeaB
[0054] (1) Enhanced expression of thmgR gene
[0055] 3-Hydroxy-3-methylglutaryl-CoA reductase encoded by hmgR gene in GA 3 In the synthesis of mevalonate, it acts as a rate-limiting enzyme in the mevalonate pathway. Since the C-terminus of 3-hydroxy-3-methylglutaryl-CoA reductase can bind to the endoplasmic reticulum membrane, the products of catalyzed synthesis trigger feedback inhibition. Using primers 23 and 24 by PCR, the gpdA promoter sequence was amplified using the plasmid pAN7-1 as a template, and the PCR reaction conditions were as follows: 98°C for 5 minutes; 98°C for 30s, 60°C for 30s, and 72°C for 30s, repeating 30 cycles; Continue at 72°C for 5 minutes.
[0056] The N-terminus of 3-hydroxy-3-methylglutaryl-CoA reductase responsible for catalysis was amplified by PCR using primers 25 and 26, using the genome of Fusarium fujikuroi CCTCC NO: M2019378 as a template For the coding region, the P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com