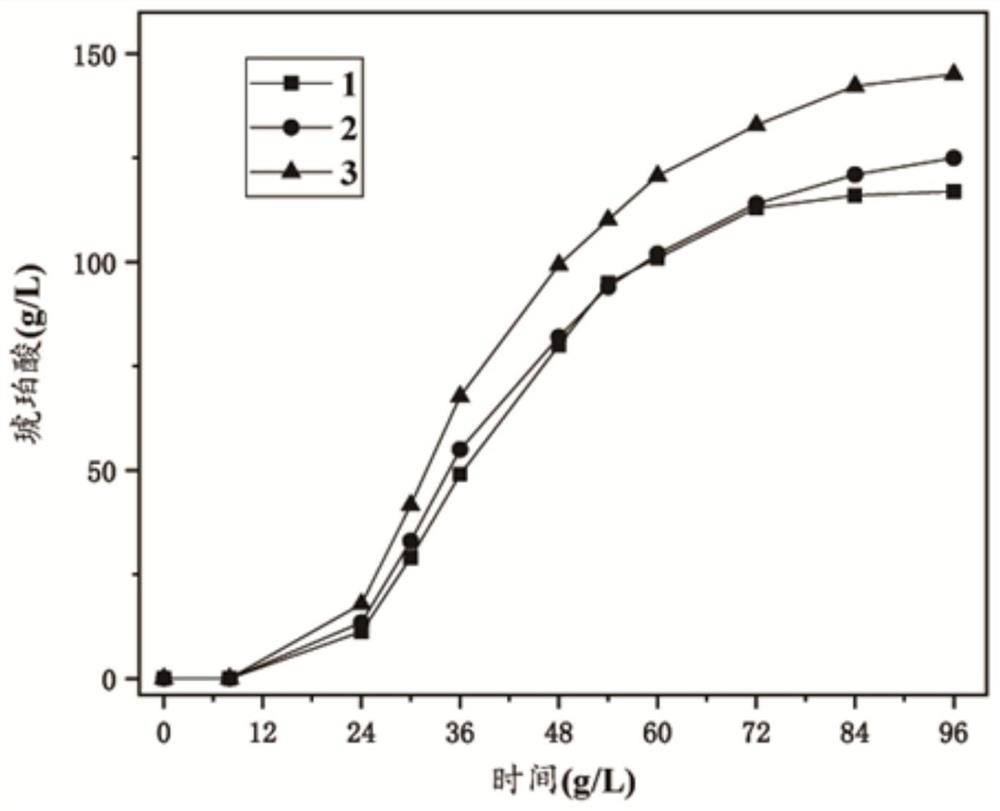
Recombinant escherichia coli with high succinic acid yield and construction method and application thereof
A technology for recombining Escherichia coli and Escherichia coli, which is applied in the field of bioengineering, can solve the problems of cofactor metabolic imbalance, metabolic imbalance, product yield and low production intensity, etc., to improve the ability of acid resistance and hyperosmotic resistance, multi-reduction force effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1E
[0055] Construction of Example 1E.coli FMME-N-5 (ΔfocA-pflB-ΔldhA-Δpta-ackA) CRISPR-Cas9 system
[0056] The Escherichia coli CRISPR-Cas9 system consists of two basic plasmids, pCas-lac and pTargetF. Plasmid pCas-lac is an episomal plasmid of Escherichia coli, which contains L-arabinose-induced expression of Red recombinase element, Cas9 protein coding gene Cas9, thermosensitive Type elements and the sgRNA used to induce and eliminate the plasmid pTargetF, the kanamycin resistance gene KanR, etc. All plasmid series containing pCas-lac derivatives need to be cultured at 30°C to ensure normal replication of the plasmid without loss. Plasmid pTargetF is an E. coli episomal plasmid containing the spectinomycin resistance gene aadA and the promoter pJ23119 for transcription of sgRNA.
[0057] According to the sequence of the target editing site, primers were designed to amplify a 20-base sequence (N20) that matched the target site, and then the sequence was cloned into the pTarget...
Embodiment 2
[0079] Example 2: Construction of E.coli FMME-N-5 (ΔfocA-pflB-ΔldhA-Δpta-ackA)-ΔfdhF-fdh1 strain
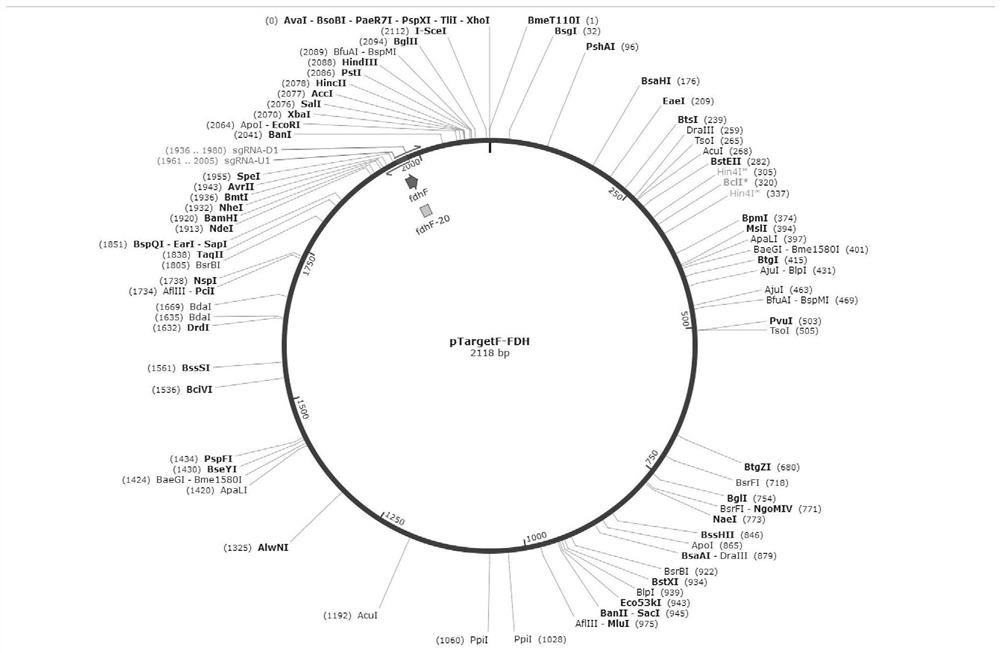
[0080] (1) Design primers according to the upstream and downstream sequences of E. coli fdhF gene. According to the Escherichia coli genome sequence published on NCBI, find the formate dehydrogenase fdhF sequence, select the cleavage site N20 (agatccgctacaaactgacg) on the fdhF gene, design the reverse amplification primers for pTargetF full plasmid PCR, and obtain the pTargetF-fdhF knockout plasmid ,Such as figure 1 As shown, wherein, the primers are as follows:
[0081] sgRNA-U1:agatccgctacaaactgacggttttagagctagaaatagcaagtt
[0082] sgRNA-D1: cgtcagtttgtagcggatctactagtattatacctaggactgagc.
[0083] (2) PCR method was used to amplify the upstream homology arm, downstream homology arm and fdh1 homology arm replaced by fdh1, and the primers were as follows:
[0084] Amplified FDH1 replaces the upstream homology arm primer
[0085] F-U1: cgttacaaccagtcagtactgaacg
[0086] F-D...
Embodiment 3
[0094] Example 3: RBS optimized expression of the outer membrane protein regulator OmpR
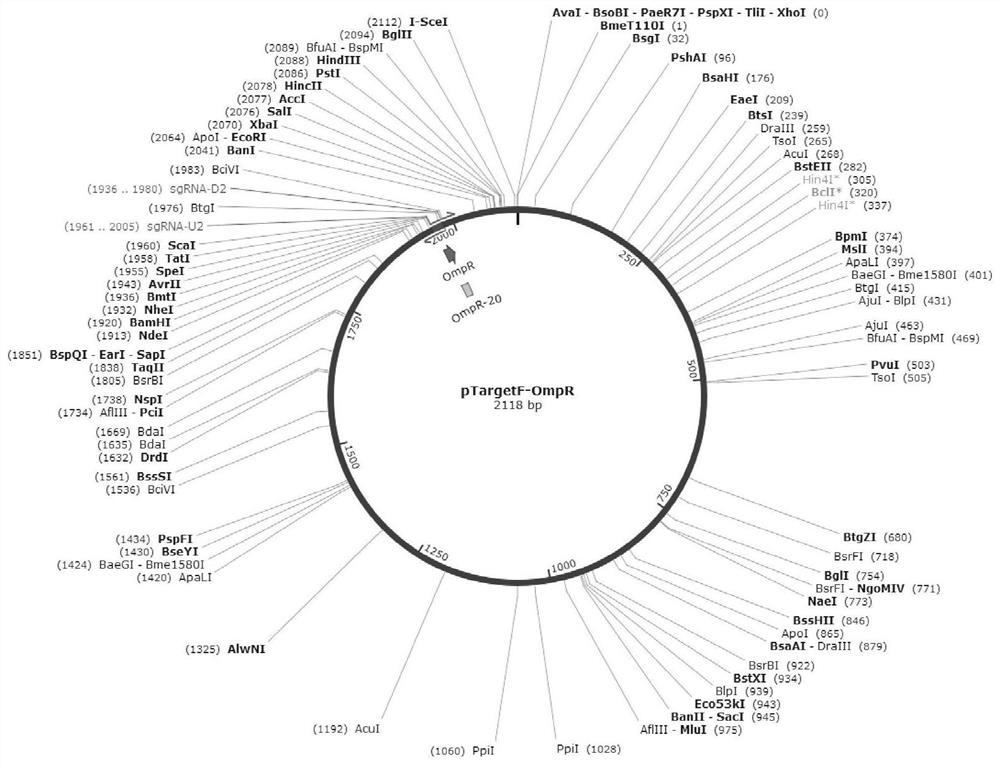
[0095] (1) Primers were designed according to the upstream and downstream sequences of the Escherichia coli outer membrane protein regulator OmpR gene. According to the Escherichia coli genome sequence published on NCBI, the sequence of the outer membrane protein regulator OmpR was found, and the cleavage site N20 (actgctggcccgtatccgtg) was selected on the gene, and the reverse amplification primers for pTargetF full plasmid PCR were designed to obtain pTargetF-OmpR knockout plasmids, such as figure 2 As shown, wherein, the primers are as follows:
[0096] sgRNA-U2: actgctggcccgtatccgtggttttagagctagaaatagcaagtt
[0097] SGRNA-D2: cacggatacgggccagcagtactagtattatacctaggactgagc.
[0098] (2) Synthesize the gene sequence J23101-RBS8-OmpR in which OmpR, RBS8 and the artificial promoter J23101 are continuous by the method of gene synthesis, and the gene sequence is shown in SEQ ID NO.4.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com