Cracking polysaccharide monooxygenase and application thereof
A polysaccharide monooxygenase and enzymatic hydrolysis technology, applied in the field of bioengineering, can solve the problems of difficult and efficient hydrolysis, unfriendly environment, high energy consumption, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
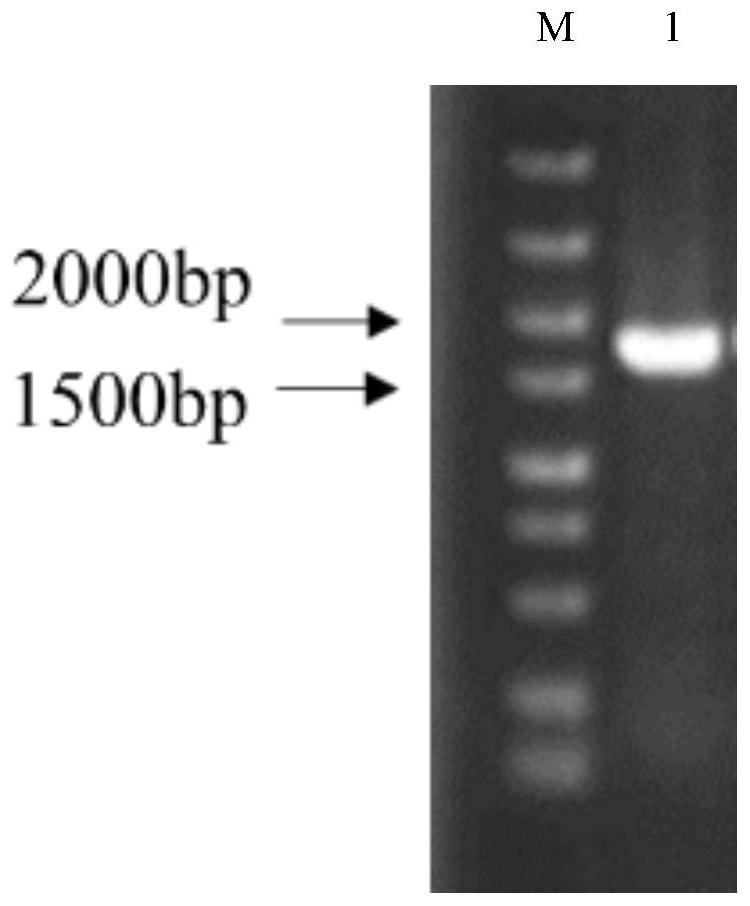
[0051] Embodiment 1: the acquisition of lytic polysaccharide monooxygenase PlLPMOCB3 target gene
[0052] (1) The target gene was synthesized by Jinweizhi Company according to SEQIDNO.2, and 5'(EcoRI) and 3'(EcoRI) restriction sites were added during the synthesis, and the gene was cloned into the vector pET-28a through 5'EcoRI and 3'EcoRI (+) Construction of recombinant plasmids. The nucleotide sequences of the upstream and downstream primers are shown below:
[0053] Upstream primer:
[0054] 5'-ATGGGTCGCGGATCCATGATTCAAAACGTTCACGTTCAA-3';
[0055] Downstream primers:
[0056] 5'-TTGTCGACGGAGCTCGAATTCTTAATGGTGGTGGTGATGATG-3';
[0057] Plasmid pET-28a(+) was linearized, and the linearized plasmid was purified by DNA Extraction Mini Kit.
[0058] (2) Use T4 DNA ligase to connect the digested vector and the target base fragment together. The ligation system is as follows: 5 μL of the target gene, 3 μL of linearized vector, 1 μL of T4 DNA ligase, and 1 μL of Buffer. Ligatio...
Embodiment 2
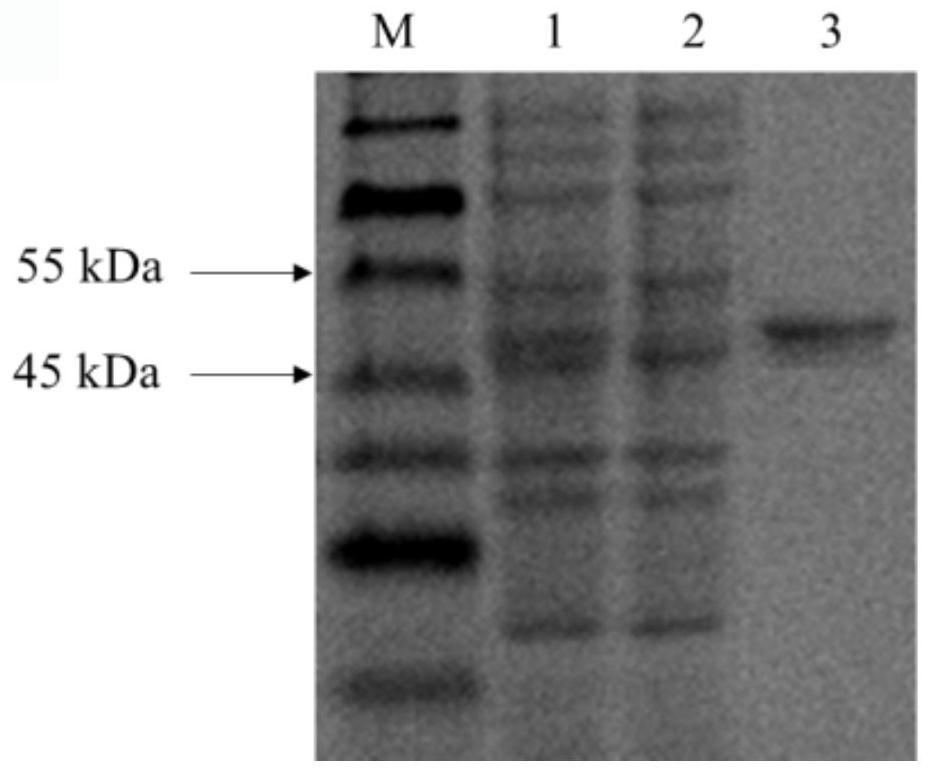
[0060] Example 2: Construction and induction expression of recombinant Escherichia coli Rosetta-CB3
[0061] (1) Transformation of Escherichia coli. Take 10 μL of the recombinant pET-28a(+) plasmid constructed in Example 1 and add it to 80 μL of L. coliRosetta competent cells, mix gently and place on ice for 30 min, heat shock in a water bath at 42°C for 90 s, and immediately ice bath for 2-5 min. Add 1 mL of LB medium without resistance, resume culture at 220 rpm and 37 °C for 1 h, centrifuge to take 100-200 μL of bacterial liquid and spread it on LB solid medium containing kanamycin and chloramphenicol resistance, invert at 37 °C Incubate overnight. Pick a single colony on the LB plate into 5 mL of LB liquid medium containing kanamycin and chloramphenicol resistance, and cultivate at 37 ° C and 220 rpm for 12 h. Using the bacterial liquid as a template, PCR was performed at 1% (w / v ) of agarose gel electrophoresis verification results, screened positive clones for verifica...
Embodiment 3
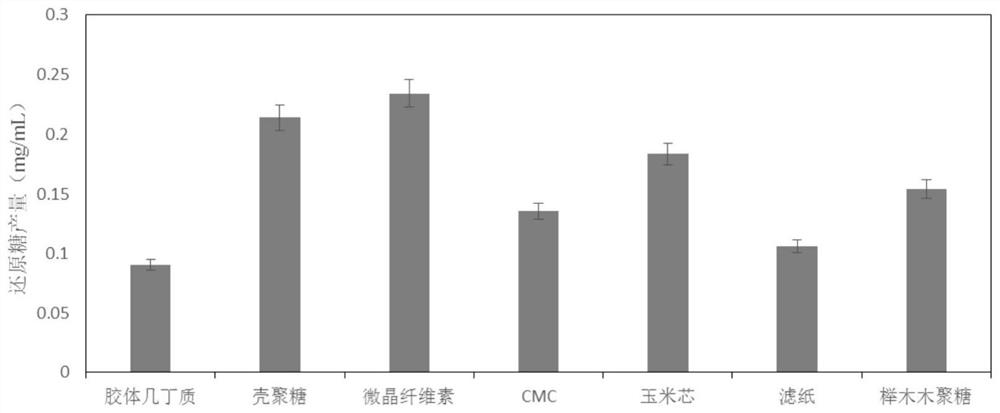
[0065] Example 3: Substrate specificity analysis of PlLPMOCB3
[0066] Take several 1.5mL centrifuge tubes, add 1% (w / v) final concentration of colloidal chitin, chitosan, microcrystalline cellulose, CMC, corncob powder (pulverized and dried), filter paper, Beech wood xylan, add purified PlLPMOCB3 to make the final concentration 1 mg / mL, add ascorbic acid to make the final concentration 1 mM, use 50 mM, pH 5.0 sodium citrate buffer solution to accurately make up the volume to 1 mL, seal with parafilm Incubate at 50°C, shake at 220rpm for 36h, heat in boiling water for 5min to terminate the reaction, centrifuge at 12000rpm for 1min to collect the supernatant, and measure the yield of reducing sugar by PAHBAH method. The reaction system without enzyme was used as a blank control, and all samples were replicated 3 times each.
[0067] The result is as image 3 As shown, PlLPMOCB3 can act on colloidal chitin, chitosan, microcrystalline cellulose, CMC, corncob, filter paper and b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com