Phytase variants
A technology of phytase and Aspergillus phytase, applied in the field of phytase variants, can solve problems such as phosphate pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 7
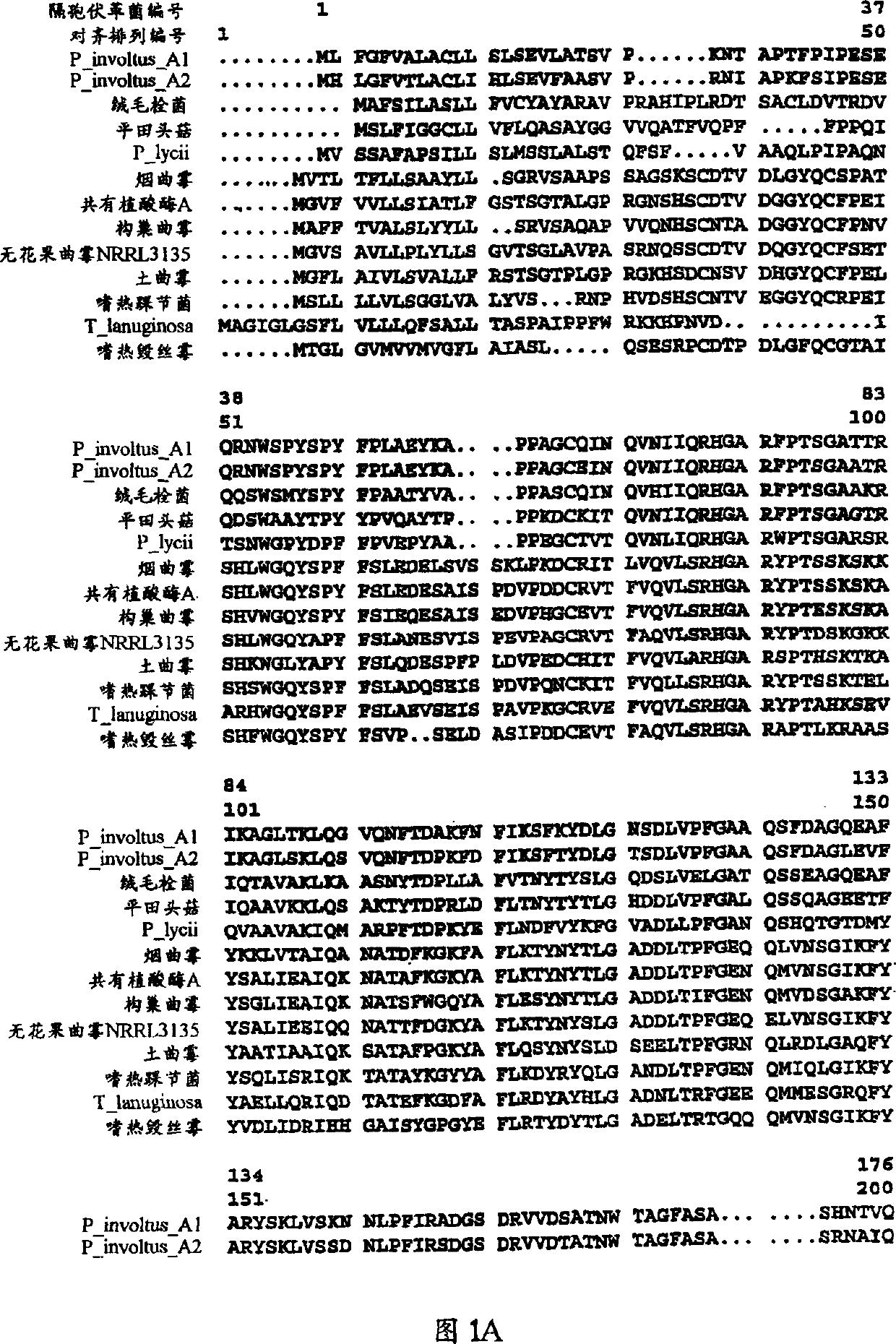
[0065] Example 7 shows an example of how to add a novel model phytase to the alignment of Figure 1 and deduce its corresponding phytase variant.
[0066]Similar to Example 7, other model phytases can be aligned and variants deduced. This is especially true for the following model phytases: the phytase of Aspergillus niger var. awamori (US Patent No. 5,830,733); the Bacillus phytase of WO 98 / 06858; the soybean phytase of WO 98 / 20139; The corn phytase of WO 98 / 05785; the Aspergillus phytase of WO 97 / 38096; the Monascus phytase of WO98 / 13480;
[0067] When a model phytase and a putative phytase variant are compared using the alignment described herein, the corresponding amino acid positions, i.e., the model phytase and the variation of the variant can be identified Loci - Simply place the corresponding pattern loci and variant loci one above the other in the alignment. An alteration is said to have occurred at a given position if the pattern amino acid at the pattern position i...
Embodiment 1
[0328] Phytase Activity Test (FYT)
[0329] Phytase activity can be measured using the following assays:
[0330] 10 μl of diluted enzyme sample (diluted in 0.1 M sodium acetate, 0.01% Tween 20, pH 5.5) was added to 5 mM sodium phytate (Sigma) in 0.1 M sodium acetate, 0.01% Tween 20, pH 5.5 250 μl (adjust pH after dissolving sodium phytate; preheat substrate) and incubate at 37° C. for 30 minutes. The reaction was terminated by adding 250 μl of 10% TCA, and added in 100 ml of molybdate reagent (will be 2 SO 4 2.5g dissolved in (NH 4 ) 6 Mo 7 o 24 4H 2 O diluted to 7.3g FeSO dissolved in 250ml) 4 500 μl to determine free phosphate. The absorbance at 750 nm of 200 µl of samples was measured in a 96-well microtiter plate. Simultaneous determination of substrate and enzyme blank control. A phosphate standard curve (0-2 mM phosphate) is also included. 1 FYT is equal to the amount of enzyme that releases 1 μmol of phosphate per minute under the given conditions.
Embodiment 2
[0332] Test for specific activity
[0333] Specific activity can be determined as follows:
[0334] A highly purified phytase sample (preliminarily checked for purity on an SDS polyacrylamide gel showing only one component present) was used.
[0335] The protein concentration in the phytase samples was determined by amino acid analysis as follows: A phytase sample was hydrolyzed in 6N HCl, 0.1% phenol for 16 hours at 110°C in an evacuated glass test tube. The obtained amino acids were quantified using an Applied Biosystems 420A amino acid analysis system in accordance with the manufacturer's instructions. From the amount of amino acids it is possible to calculate the total mass - and thus concentration - of protein in the hydrolyzed sample.
[0336] Activity was determined in units of FYT. One FYT is equal to the amount of enzyme that releases 1 micromole of inorganic phosphate per minute from phytate (5 mM phytate) at pH 5.5 at 37°C; the assay method is described eg in Exa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com