Immunity regulating type Eimeria tenella DNA vaccine for preventing and treating chicken coccidiosis
A DNA vaccine, Eimeria technology, applied in the field of DNA vaccines, can solve the problems of high cost, not fully applicable to laying hens and broilers, and poor stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Preparation of embodiment 1.pEtK2 gene:
[0043] 1. Synthesize primers P1 and P2, the sequences are:
[0044] The following primers were designed according to the published nucleotide sequence of pEtK2 (Genebank accession number AY389513):
[0045] P1: 5'-AA GGATCC CCAGCAGCGTCCAA-3′
[0046] P2: 5'-GAA GAATTC CGCTGCGGTATCTCCG-3'
[0047] The underlined parts are the introduced restriction sites BamHI and EcoRI, respectively; the boxed part is the initiation codon.
[0048] 2. Extraction of coccidia total RNA:
[0049] Take 0.1 g (about 107) of pure sporulated oocysts of E. tenella, put them in a glass homogenizer, add 1 ml of Trizol reagent, grind for 5 minutes to rupture the oocyst wall, and extract the total sporozoite RNA in one step. The specific steps are as follows: put the homogenized solution into a 1.5ml eppendorf tube, add 0.2mL of 24:1 chloroform / isoamyl alcohol mixture, shake the sample vigorously for 15s, put it on ice for 5min, centrifuge at 1200...
Embodiment 2
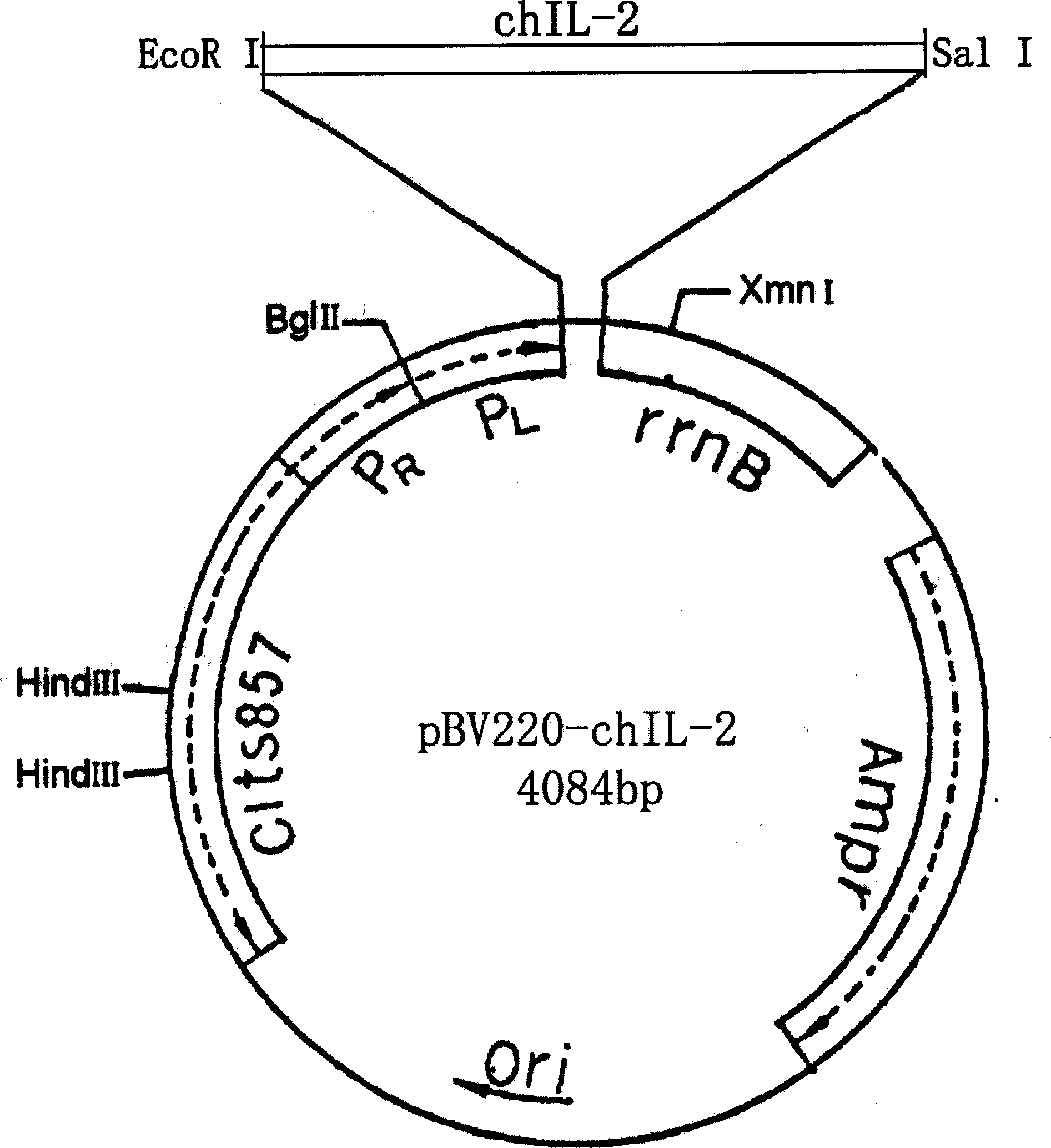
[0074] Example 2. Preparation of chIL-2 gene:
[0075] 1. Synthesize primers P3 and P4, the sequences are:
[0076] P3: 5'-CTA GAATTC CCTACCCTCGAT TGCAAAGTACTGATCT-3'
[0077] P4: 5'-TTA GTC GAC TTGCAGATATCTCCAAAAGTT-3'
[0078] The underlined part is the introduced enzyme cutting site EcoRI and SalI; the square part is the start codon; the italic letter part is the coagulation factor X a The specific hydrolysis sequence encodes a nucleotide sequence (such as the nucleotide sequence shown at positions 1465-1482 in SEQ ID NO: 1).
[0079] 2. PCR amplification of chIL-2 gene
[0080] Add the following components to a thin-walled PCR tube for PCR amplification:
[0081] Plasmid pBV220-chIL-2 (50ng / μl) 1.0μl
[0082] 2.5mmol / L dNTP 4.0μl
[0083] P3 (50pmol) 1.0μl
[0084] P4 (50pmol) 1.0μl
[0085] 10×PCR buffer 5.0μl
[0086] 25mmol / L MgCl 2 23.0μl
[0087] Taq DNA polymerase (5U / μl) 0.5μl
[0088] wxya 2 O 34.5 μl
[0089] Total 50.0μl ...
Embodiment 3
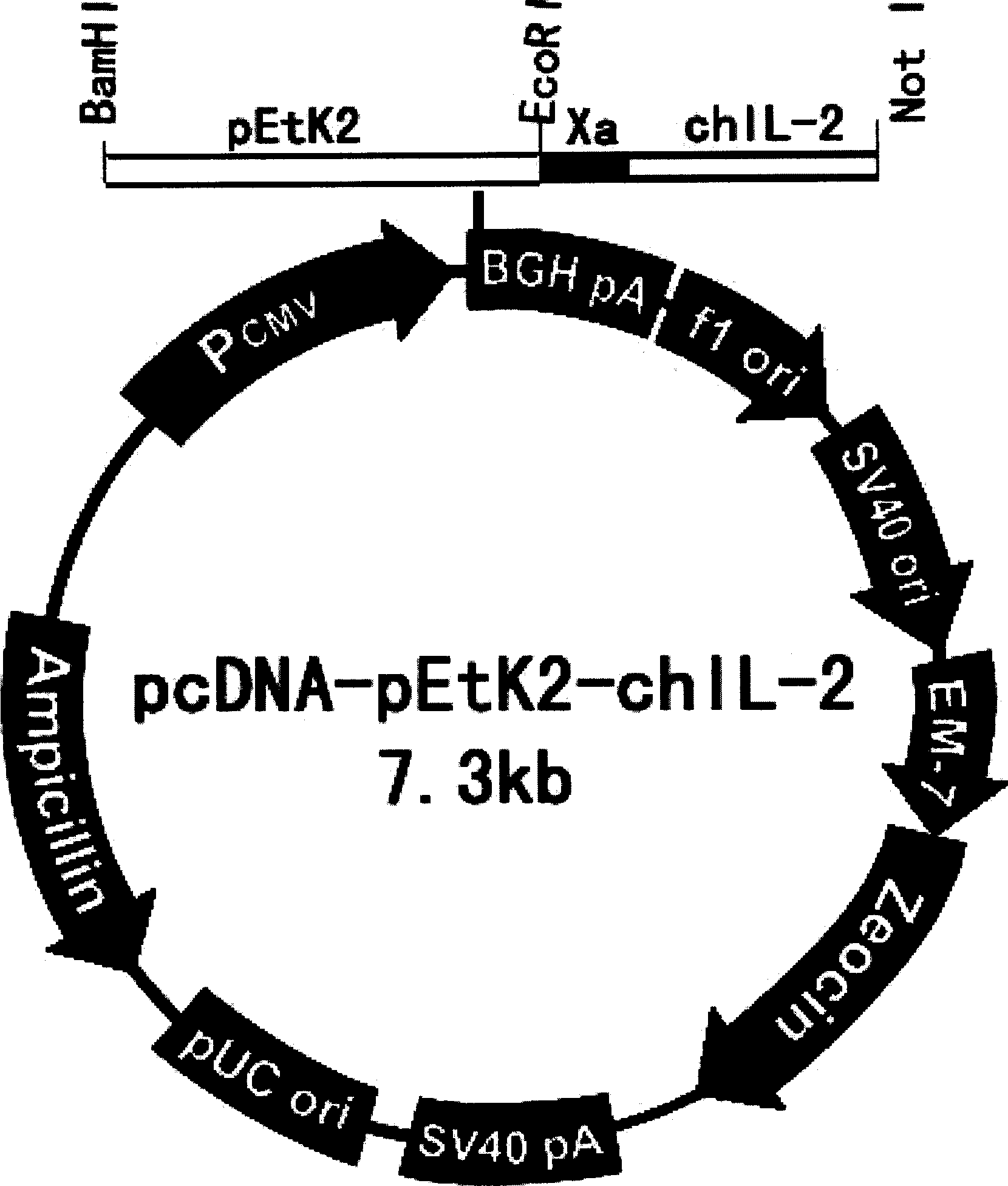
[0093] The preparation of embodiment 3.DNA vaccine
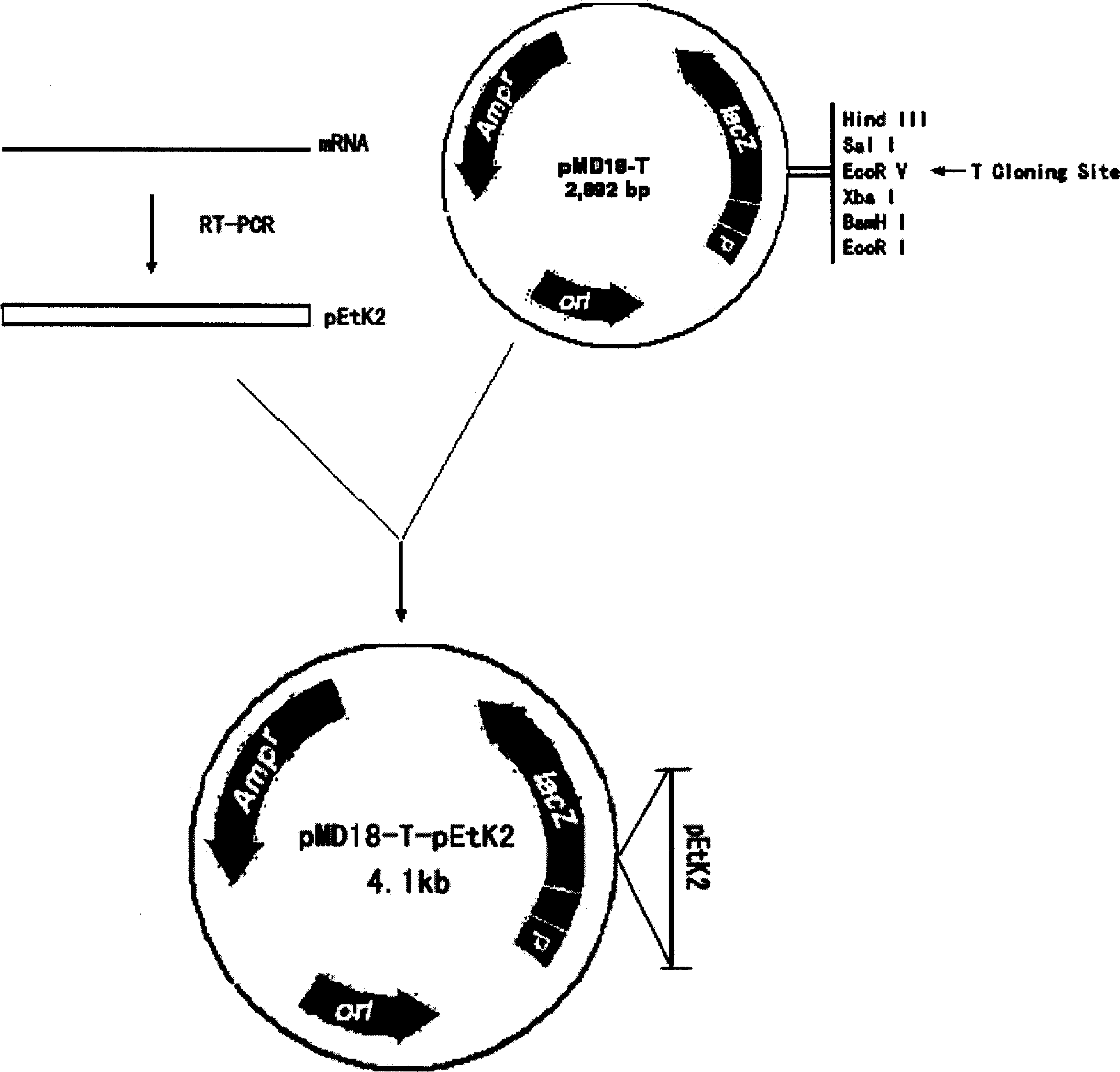
[0094] 1. Construction of recombinant plasmid pMD18-T-pEtK2 (see image 3 )
[0095] Get 50 μ l of the PCR product obtained in Example 1, electrophoresis on 1% agarose gel, cut out the agarose gel at the band of interest under ultraviolet light, reclaim and purify the fragment of interest with the gel recovery kit of Dalian Bao Biological Company, method Refer to the instruction manual. Take the purified PCR product and connect it with the pMD18-T vector linearized after digestion with BamHI and EcoRI. The reaction system is as follows:
[0096] PCR recovery product 4.0μl
[0097] pMD18-T vector 1.0μl
[0098] Ligation solution I 5.0μl
[0099] Total volume 10.0μl
[0100] The above components were mixed in a thin-walled eppendorf tube and connected overnight at 4°C. The ligation product was transformed into competent Escherichia coli JM109, the plasmid was extracted, and identified by double enzyme digestion and se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com