Automated hybridization/imaging device for fluorescent multiplex DNA sequencing
a hybridization/imaging device and fluorescent multiplex technology, applied in the field of automatic hybridization/imaging device for fluorescent multiplex dna sequencing, can solve the problems of hazardous and unstable acquisition of sequence data, inability to achieve high-resolution direct imaging of radioactive signals, and inability to achieve straight-forward removal. , to achieve the effect of easy removal
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
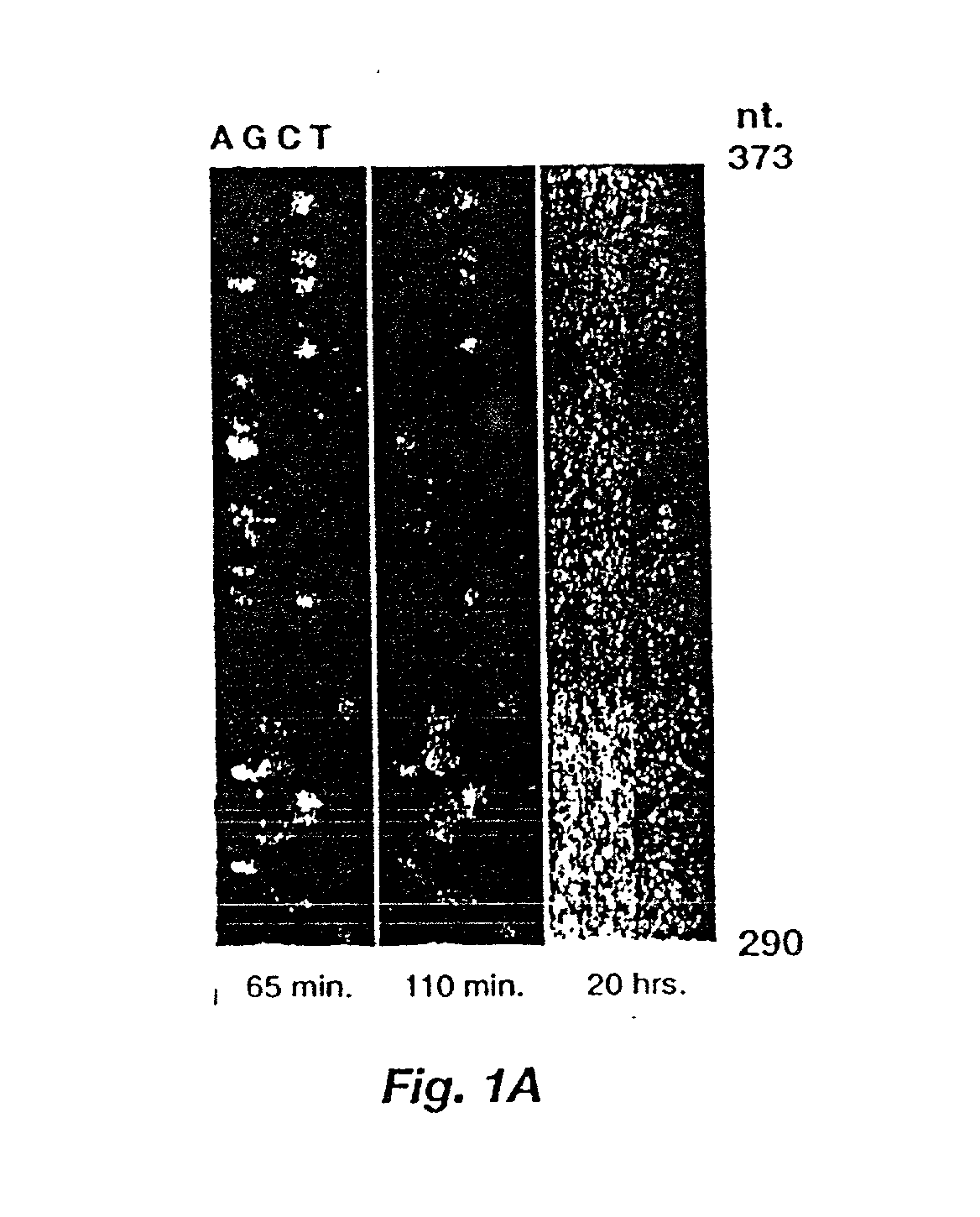
[0042] The membrane from Example 1 was placed in an automated imaging hybridization chamber, to be described momentarily, where it was probed with a biotinylated oligonucleotide complementary to positions 80 to 110 of the ladder, and then treated with a streptavidin-alkaline phosphatase conjugate. Complete cycle time was 4.5 hours. The rotation of the inner cylinder within the hybridization chamber device was set to sweep a bead of fluid across the convex surface of the membrane, by movement of the membrane through the fluid, at approximately 20 second intervals. Hybridization volumes were 50 ml total, and wash volumes were 100 ml each. Unbound probe was removed by 8 washes with phosphate buffered saline (PBS) containing 5% SDS. Then a 1 / 5000 dilution of streptavidin-alkaline phosphatase (Boehringer Mannheim, 1000 U / ml) was applied in a total volume of 40 ml of PBS, 5% SDS. The enzyme conjugate was allowed to bind for 45 minutes. Then, unbound conjugate was removed by 1 wash with PB...
example 3
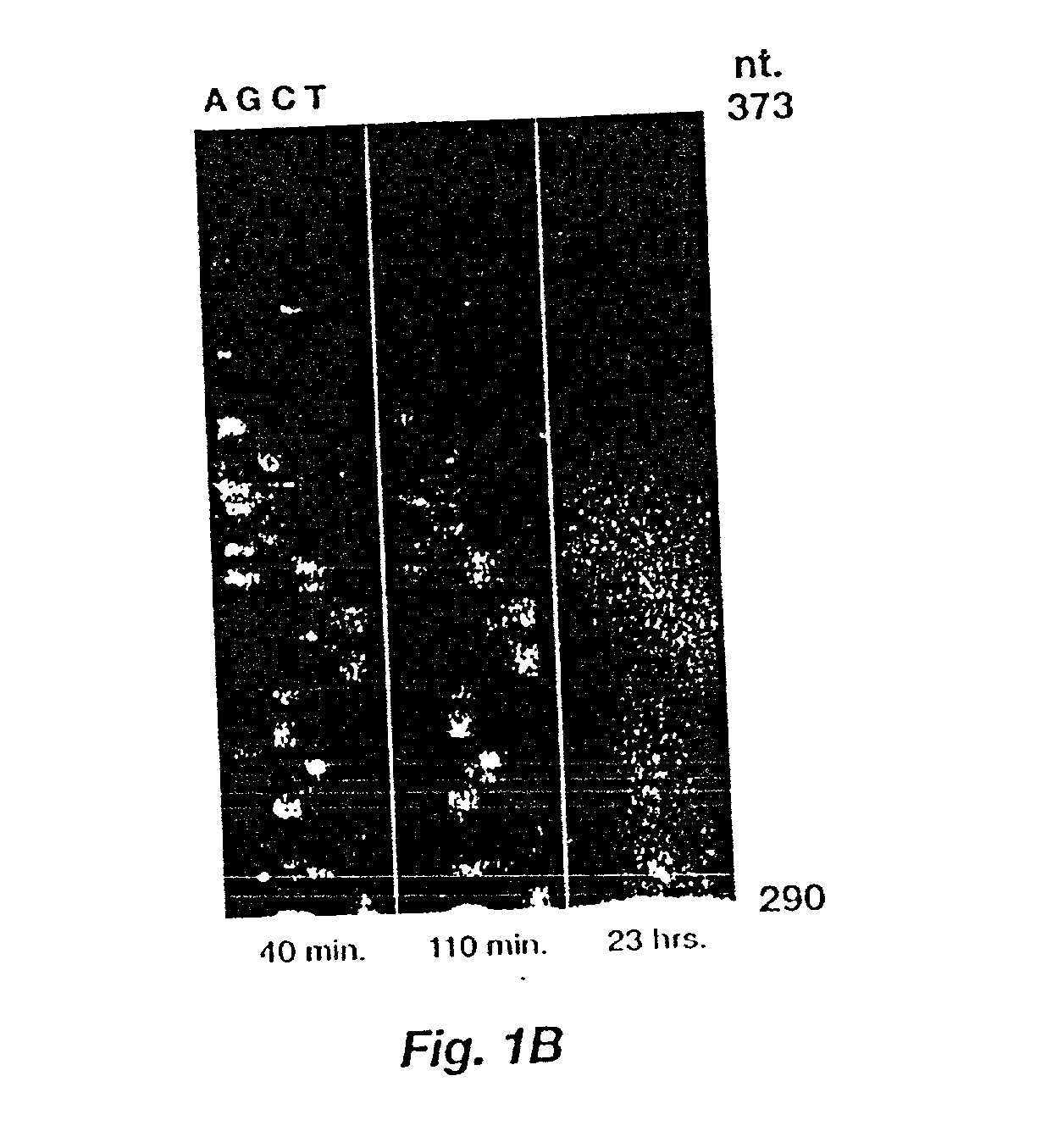
[0043] The membrane from Example 2 was cut so that each of the three substrates could be applied to a sequencing ladder. The enzymatic reaction was started by addition of 1 ml of a fluorogenic alkaline phosphatase substrate for every 300 cm.sup.2 of membrane. The stock solutions were: MUFP-50 .mu.g / ml in 0.1 M diethanolamine, pH 10; 5MFP-50 .mu.g / ml in 0.1 M diethanolamine, pH 10; and BBTP-600 .mu.g / ml in 2.4 mM diethanolamine, pH 10. The substrate solution was applied as an even coat on the membrane.
[0044] In this example, the membranes were imaged outside the chamber, thus membranes were then placed on glass plates and wrapped with transparent plastic film. Fluorescence emission was excited by a 458 nm line of an argon ion laser (Lexel model 96) for 5MFP and BBTP or a long wave UV mercury lamp for MUFP. Laser light was passed through a lens to widen the beam distribution and through a 450 nm, 40 nm bandwidth bandpass filter (Melles Griot). Images were obtained by a cryogenically c...
example 4
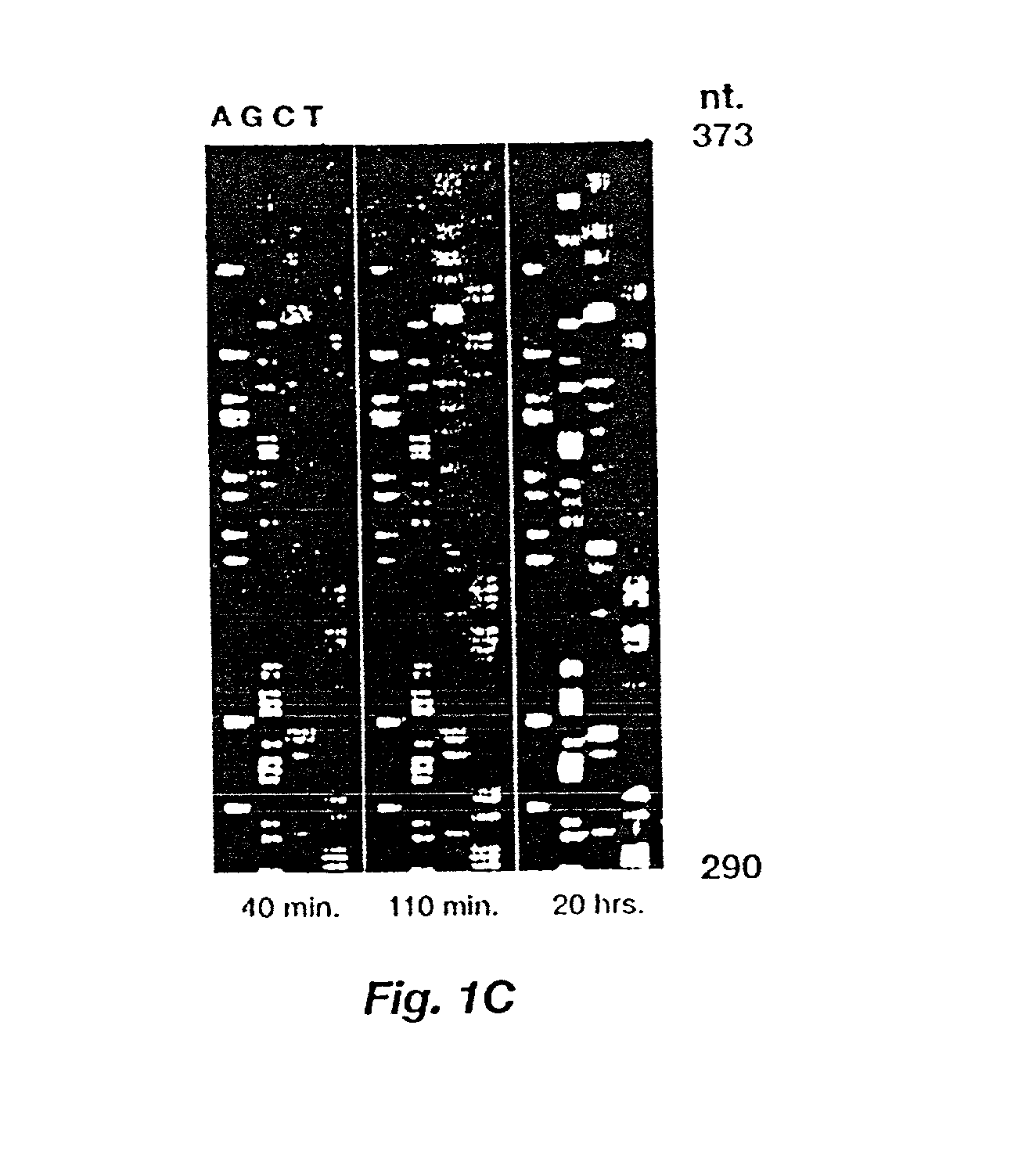
[0062] A full length image of the BBTP membrane, 40 minutes after addition of substrate, shown in FIG. 1C was acquired with a CCD camera operating in TDI mode and was processed using the automatic gel reading software of serial no. 07 / 978,915 modified to accept a positive fluorescent image rather than a negative one as is usual with imaging on X-ray film. FIG. 5 shows a portion of the unprocessed one-dimensional traces, obtained by averaging horizontally across several pixels at the lane centers. The BBTP membrane was up-sampled by a factor of two to produce a 300 dots-per-inch image expected by the gel reader. Lanes were found manually using a Macintosh program, and the data shown were produced by averaging the value of 15 pixels per row around the lane centers. The individual traces have not been processed in FIG. 5, but the traces have been slightly shifted relative to one another to obtain better alignment.
[0063] The automated reader yielded reasonably accurate sequence as far a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com