Gene chip and test kit for detecting important pathogenic bacteria in aquatic products
A gene chip and pathogenic bacteria technology, applied in the field of gene chips and kits for detecting important pathogenic bacteria in aquatic products, can solve problems such as identification difficulties, and achieve the effects of simple operation, strong repeatability and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Design and preparation of probes
[0035] 1. Sequence acquisition:
[0036] (1) Acquisition of bacterial 16S rDNA sequences: all 16S rDNA sequences of ten species of bacteria were downloaded from the GenBank public database.
[0037] (2) Acquisition of 16S-23S rDNA interval sequence: download from the GenBank public database to obtain Salmonella, Vibrio parahaemolyticus, Vibrio cholerae, Listeria monocytogenes, Staphylococcus aureus, Streptococcus pyogenes, and their All 16S-23S rDNA interregion sequences of closely related bacteria.
[0038] In order to meet the needs of intra-species conservation and inter-species specificity in target sequence analysis, 48 strains of Proteus (including all 4 species of this genus) were selected for the genus Proteus with few ITS resources: 22 strains of Proteus mirabilis, 14 strains of Proteus common strains, 10 strains of Proteobacterium panethii, and 2 strains of Proteus myxogenes) were sequenced in the interregion,...
Embodiment 2
[0051] Example 2 Design and preparation of primers
[0052] 1. Sequence acquisition: the same as the sequence of the designed probe.
[0053] 2. Design primers:
[0054] (1) Design of primers for amplifying the interregional sequence: After comparing the 16S rDNA of ten species of bacteria downloaded from the public database NCBI with the sequence alignment software Glustal X, a 16S rDNA conserved sequence close to the interregional region was selected as the upstream primer , the length conforms to T m Value 50°C±5°C, length 17bp±2bp, Hairpin: NONE, Dimer: NONE, False Priming: NONE, CrossDimer: NONE, and contains bacterial universal probe sequence.
[0055] (2) Design of primers for amplifying the ipaH gene sequence: the above-mentioned ipaH gene sequences of the four species of Shigella downloaded from the GenBank public database were compared with the sequence alignment software Glustal X to find the conserved segment of the gene, The conserved segment was imported int...
Embodiment 3
[0061] Example 3 Gene Chip Preparation——Chip Spotting
[0062] 1. Dissolving probes: the probes synthesized in Example 1 were respectively dissolved in 50% DMSO solution, and diluted so that the final concentration of the probes reached 1 μg / μl.
[0063] 2. Adding plate: Add the dissolved probe to the corresponding position of the 384-well plate, 10 μl per well.
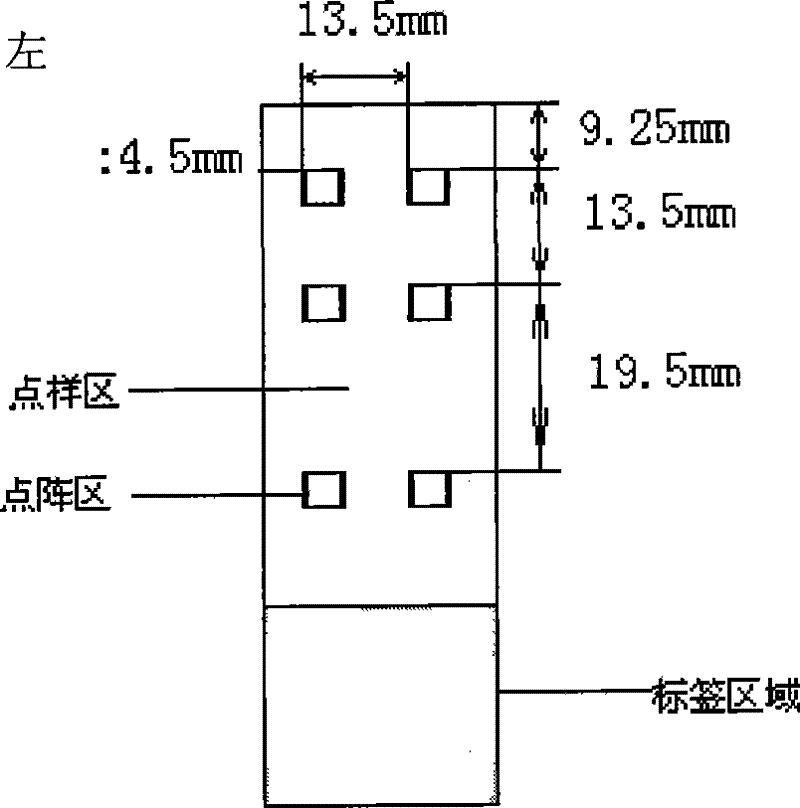
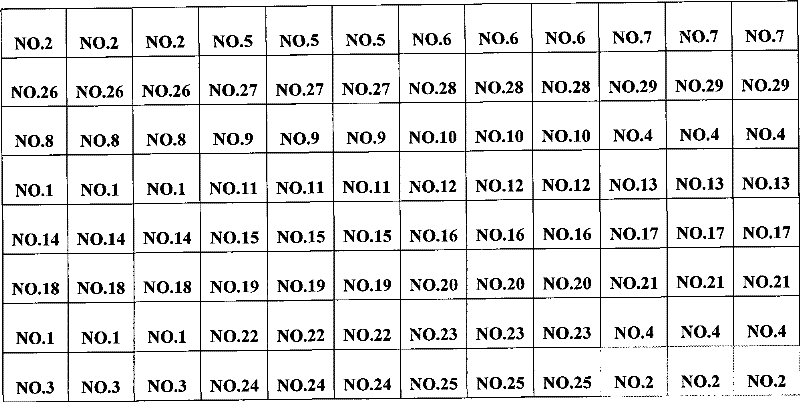
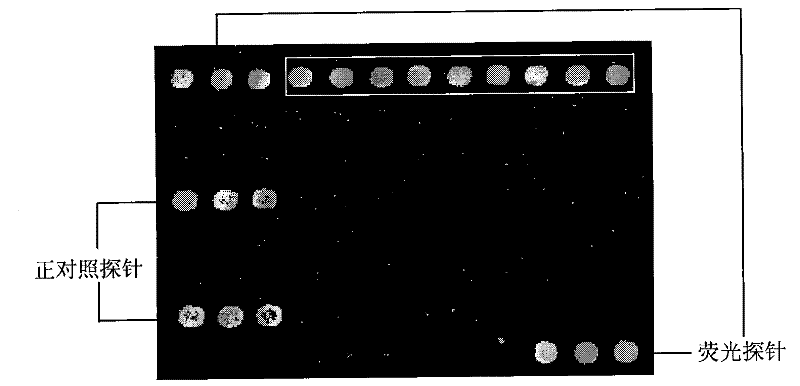
[0064] 3. Spotting: as figure 1 The shown 57.5mm × 25.5mm × 1mm (length × width × height) of the clean formaldehyde slide (CapitalBio Corp., China) was placed on the stage of the chip spotting instrument (Spotarray72), using SpotArray's Control software (Telechem smp3 stealty pin), run the program, press figure 2The arrangement shown is spotted on the aldehydeized glass slide in the spotting area of 4.5 mm×4.5 mm to form a medium-low density DNA micro-array, and the array arrangement rules in the six dot matrix areas on the glass slide are the same. The size of the dot matrix area is 3mm×2mm, the dot pitch in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com