Sucrose isomerase gene and high-efficiency expression method thereof
A technology of molasses and isomaltulose, which is applied in the fields of genetic engineering and enzyme engineering, and achieves the effects of short induction time, high expression, convenient and cheap use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Extraction of total DNA of Erwinia rhapontici NX-5.
[0032] E.rhapontici NX-5 strain (CCTCC NO: 2222) was cultured in SB liquid medium (peptone 10g / L, yeast extract 5g / L, NaCl 10g / L) for 12h, centrifuged at 8000r / min to collect the bacteria, sterile water Wash, collect the precipitate and suspend it in 500μL Tris-EDTA (Tris-hydroxymethylaminomethane-ethylenediaminetetraacetic acid) buffer, add 5μL RNase, incubate at 37°C for 30min, add 30μL 10% SDS (sodium dodecyl sulfate) And 15μL of proteinase K, incubate at 37℃ for 60min, add 100μL of 5M NaCl solution and 80μL CTAB (hexadecyltrimethylammonium bromide), incubate at 65℃ for 20min, use 700μL of phenol: chloroform: isoamyl alcohol volume The ratio of 25:24:1 mixed solution extraction, 10000r / min centrifugation, the supernatant with 700μL chloroform: isoamyl alcohol volume ratio 24:1 mixed solvent extraction, 10000r / min centrifugation, the supernatant and 1400μL of ice Mix with amyl alcohol, precipitate at -20°C...
Embodiment 2
[0033] Example 2: Cloning of SIase encoding gene.
[0034] Using the E.rhapontici NX-5 total DNA obtained in Example 1 as a template, the following nucleotide sequences were used as primers:
[0035] Primer 1: TT AAGCTT CCATGG ATTCTCAAGGATT, respectively introduce HindIII, Nco I restriction sites (underlined part).
[0036] Primer 2: GTAAATATTTGAATTAGGC GAGCTC CCTAGG TT, respectively introduce BamH I and Xho I double restriction sites (underlined part).
[0037] PCR reaction was carried out in a 20μL system: 10×PCR buffer 2μL, 2.5mmol / L dNTP 2μL, 10μmol / L primer 1 and primer 2 each 2μL, template DNA 2μL, 25mmol / LMgCl 2 2μL, TaqDNA polymerase 1μL, add double distilled water to 20μL.
[0038] PCR reaction conditions: denaturation at 95°C for 3min, then start cycling, (94°C 30s; 45°C 30s; 72°C 2min; 72°C 5min), a total of 25 cycles, amplify a 1800bp PCR fragment, cut the gel and recover the fragment After double digestion with HindIII and BamH I, the DNA fragment purification kit wa...
Embodiment 3
[0039] Example 3: Construction of pal I gene on an expression vector.
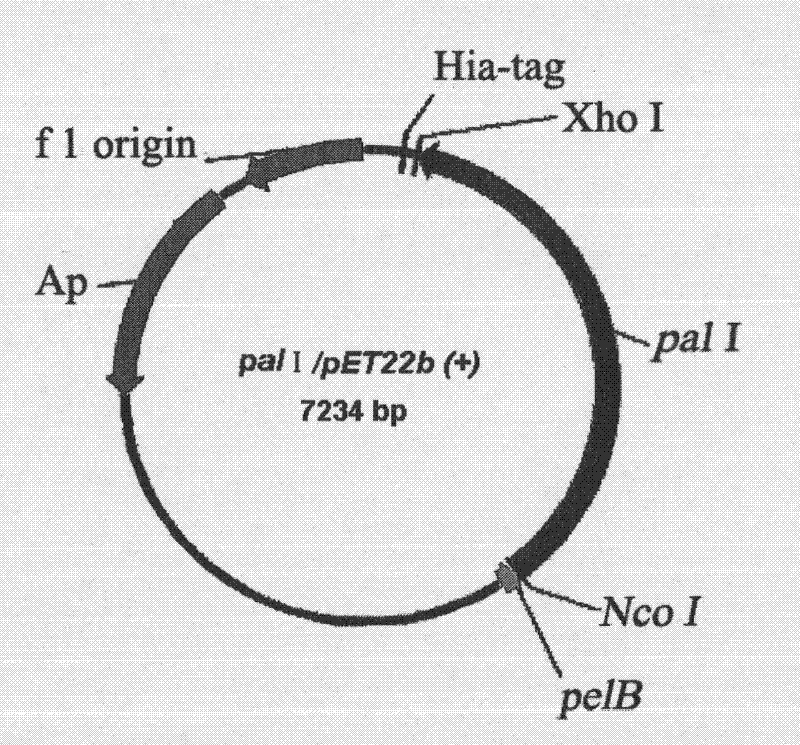
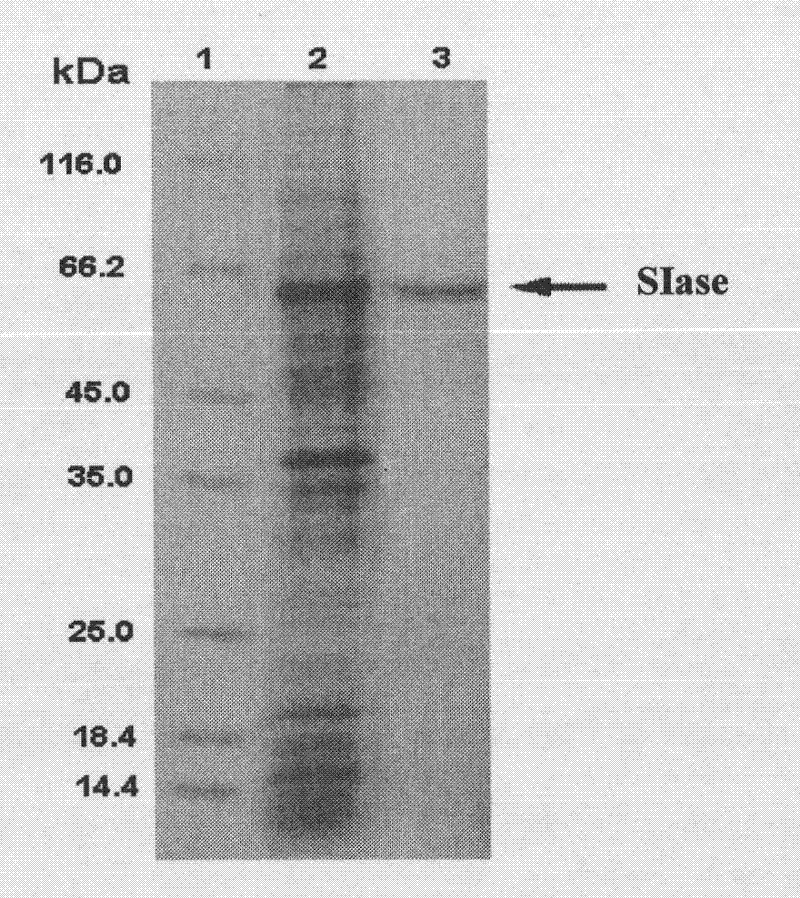
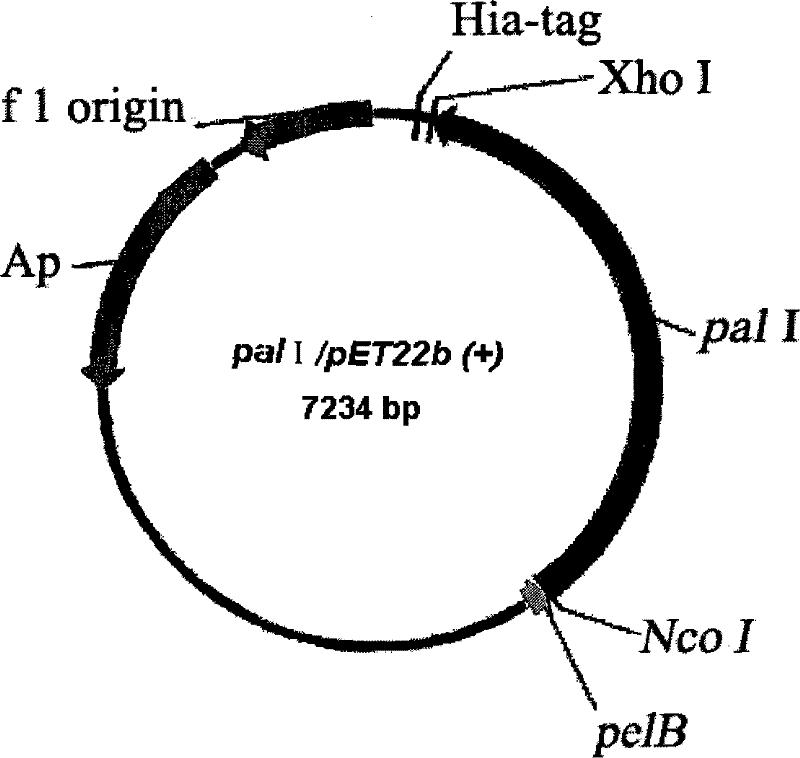
[0040] The plasmid used to construct the expression vector is pET22b(+), with pelB signal peptide and His-tag tag. The pET22b(+) plasmid and pal I gene were digested with Xho I and Nco I by double restriction enzymes. After the gel was cut to recover pal I, the pal I was ligated overnight at 16°C with T4 ligase. The ligation product was transformed into E.coli BL21(DE3) competent cells. Incubate overnight at 37°C, select the transformants for liquid culture in 100 μg / mL ampicillin LB, and then extract the plasmid to obtain the enriched pal I / pET22b(+) plasmid ( figure 1 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com