Method for extracting DNA of algae chloroplast
An extraction method and chloroplast technology, which are applied in the field of plant genetic engineering, can solve the problems of low purity of chloroplast organelles, reduced yield of chloroplast DNA, and low yield of chloroplast organelles, and achieve simple and fast extraction methods, saving operation time and high yield high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] 1) Raw material preparation: select fresh wakame algae, wash and drain, weigh 20g of algae, add 150ml of buffer A, and use a tissue homogenizer at 0-4°C to evenly grind the algae liquid;
[0033] 2) The homogenized algae liquid was filtered with 60-mesh nylon cloth, and the filtrate was centrifuged at 4,000×g for 10 minutes to obtain the precipitate;
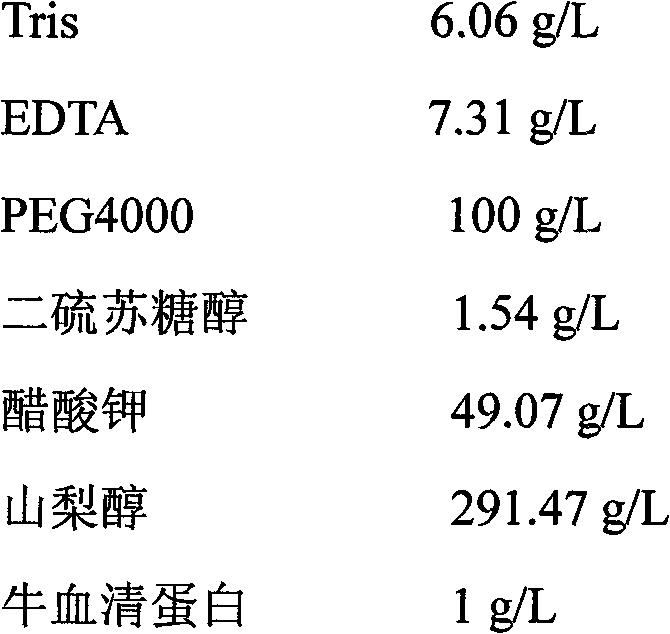
[0034] 3) Add 15ml of buffer B to the collected centrifuged precipitate at 65°C for 1 hour in a water bath;
[0035] 4) The lysate was treated with phenol-chloroform-isoamyl alcohol (volume ratio 25:24:1) and chloroform-isoamyl alcohol (volume ratio 24:1) to remove denatured proteins, and CsCl (final concentration 1.45g / ml) and Hoechst 33258 (final concentration 10μg / ml), centrifuge at 380,000×g for 8h;
[0036] 5) Use a syringe to quickly remove the band containing chloroplast DNA under ultraviolet light irradiation, and remove Hoechst 33258 with isopropanol saturated with 1M NaCl;
[0037] 6) Add 3M sodium acetate, do...
Embodiment 2
[0047] 1) Raw material preparation: select fresh Ulva algae bodies, wash and drain, weigh 25g of algae bodies, add 150ml of buffer A, and use a tissue homogenizer at 0-4°C to evenly grind them into algae liquid;
[0048] 2) The homogenized algae liquid was filtered with 60-mesh nylon cloth, and the filtrate was centrifuged at 4,000×g for 10 minutes to obtain the precipitate;
[0049] 3) Add 20ml of buffer B to the collected centrifuged precipitate, at 65°C, in a water bath for 1.5h;
[0050] 4) The lysate was treated with phenol-chloroform-isoamyl alcohol (volume ratio 25:24:1) and chloroform-isoamyl alcohol (volume ratio 24:1) to remove denatured proteins, and CsCl (final concentration 1.35g / ml) and Hoechst 33258 (final concentration 10μg / ml), centrifuge at 380,000×g for 8h;
[0051] 5) Use a syringe to quickly remove the band containing chloroplast DNA under ultraviolet light irradiation, and remove Hoechst 33258 with isopropanol saturated with 1M NaCl;
[0052] 6) Add 3M ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com