Method for expressing staphyloccocus aureus alpha-acetolactate decarboxylase by utilizing recombinant bacillus subtilis efficiently
A technology of acetolactate decarboxylase and Bacillus subtilis, which is applied in the field of genetic engineering and enzyme engineering, to shorten the aging cycle and improve production efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
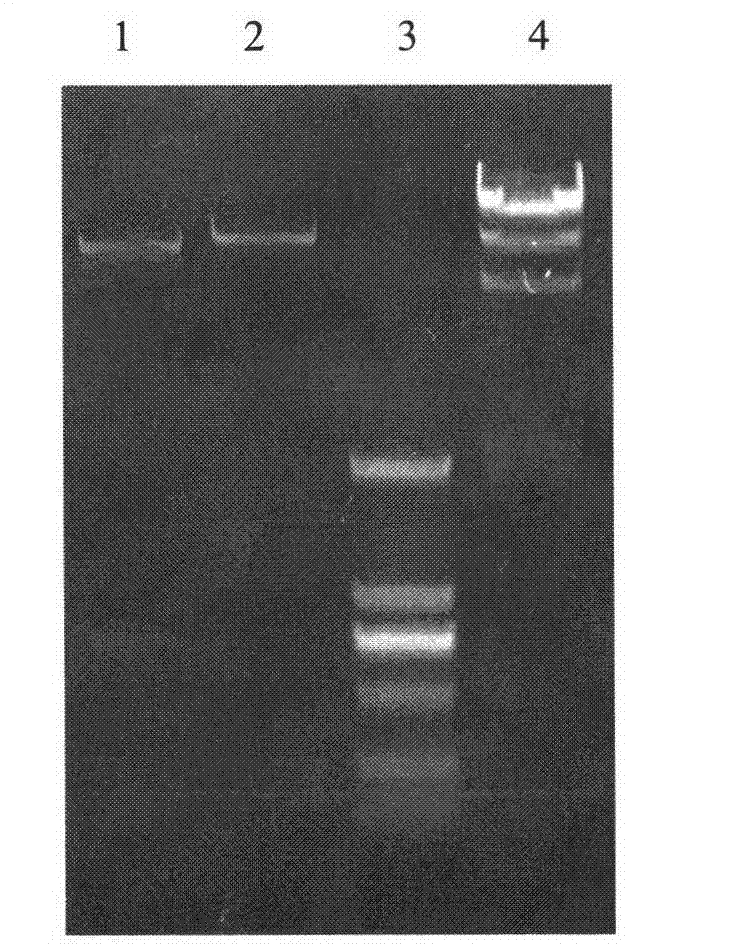
[0022] Embodiment 1: Construction and transformation of recombinant plasmid pMA5-saald
[0023] [1] Extract chromosomal DNA from Staphylococcus aureus as a template, the extraction method is as follows: use an inoculation loop to pick a single colony from a fresh Staphylococcus aureus plate and suspend it in 0.5mL sterilized double distilled water, boil it for 5 -10min, centrifuge at 8000r / min for 10min, take the supernatant into a clean 1.5mL centrifuge tube, and store in a -20°C refrigerator for later use.
[0024] [2] Using the total DNA of Staphylococcus aureus as a template, use the primers provided in Example 1 for PCR amplification. The amplification conditions are: 94°C pre-denaturation, 5min, one cycle; 94°C denaturation, 1min, 56°C annealing, 1min, 72°C extension, 1min30s, 30 cycles; 72°C, 10min, one cycle; 15°C, 10min, one cycle. PCR amplification system: template (Staphylococcus aureus chromosomal DNA) 2 μL, upstream and downstream primers 0.5 μL each, dNTP Mix 4 ...
Embodiment 2
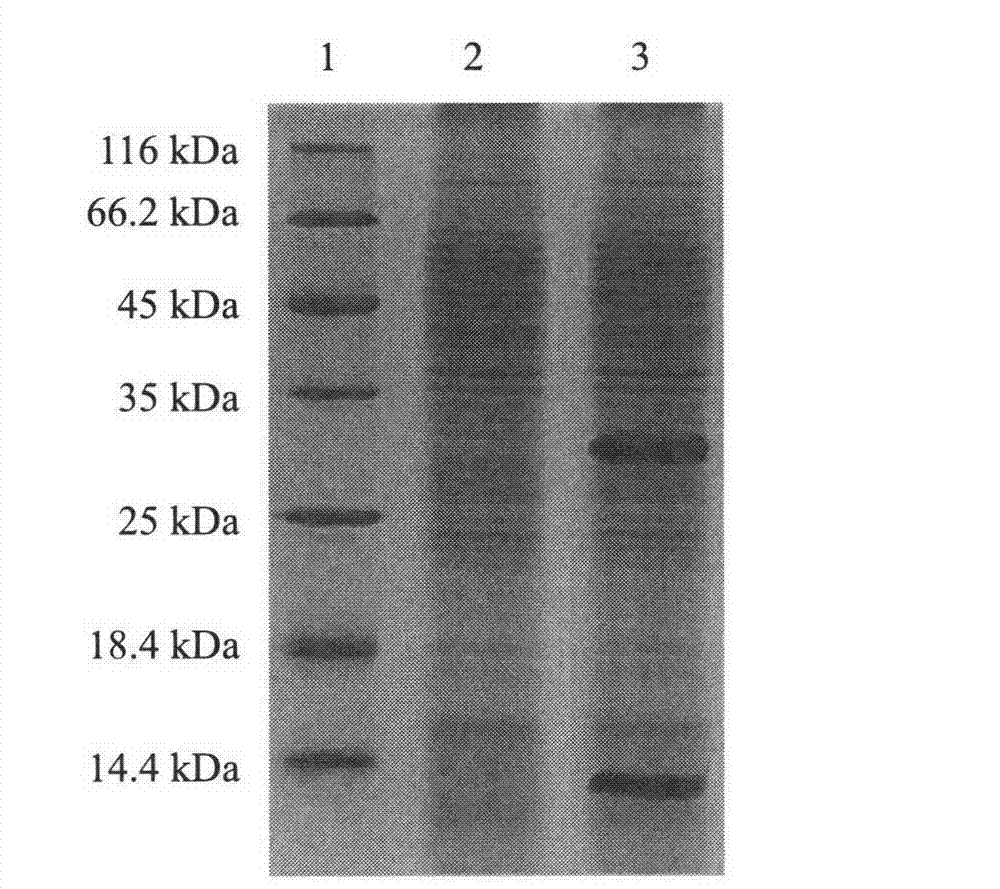
[0028] Example 2: Expression of α-acetolactate decarboxylase and assay of enzyme activity
[0029] [1] Inoculate the recombinant bacterium pMA5-saald / B.subtilis WB600 constructed in Example 1[5], the starting bacterium S.aureus and the host bacterium B.subtilis WB600 into 10 mL of liquid LB medium containing kanamycin, respectively medium, 37°C shaker at 160r / min shaking culture overnight, the next day transfer according to 1% inoculum size, culture at 37°C for 24h, take 1mL of the obtained bacterial solution in a 1.5mL centrifuge tube, centrifuge at 8000r / min for 2min, pour off the above Add 100 μL Tris-HCl buffer solution of pH 6.8 to mix the cells, add 30 μL protein gel loading buffer, bathe in boiling water for 30 minutes to denature the protein, centrifuge at 8000r / min for 10 minutes, and take the supernatant for SDS-PAGE detection , wherein SDS-PAGE uses 5% separating gel and 15% stacking gel, adopts discontinuous vertical electrophoresis, stains with Coomassie brilliant...
Embodiment 3
[0031] Embodiment 3: the fermentation of recombinant bacterium pMA5-saald / B.subtilis WB600
[0032] After the recombinant bacterium pMA5-saald / B.subtilisWB600 constructed in Example 1 [5] was activated and cultivated through LB medium, the fermentation medium was inoculated according to an inoculum size of 4% (soybean peptone 10g / L, corn steep liquor 15g / L, urea 3g / L, glucose 50g / L, k 2 HPO 4 ·3H 2 O 1.7g / L, kH 2 PO 4 2.3g / L, MgSO 4 ·7H 2 (0.75g / L, NaCl 5g / L, pH 6.8), 37 DEG C of shaker 160r / min culture 48h, finish fermentation, carry out the mensuration of enzyme activity according to the method described in embodiment 2 [2] then, its extracellular, The intracellular enzyme activities were 28300U / L and 38200U / L respectively, and the total enzyme activity reached 66500U / L.
[0033]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com