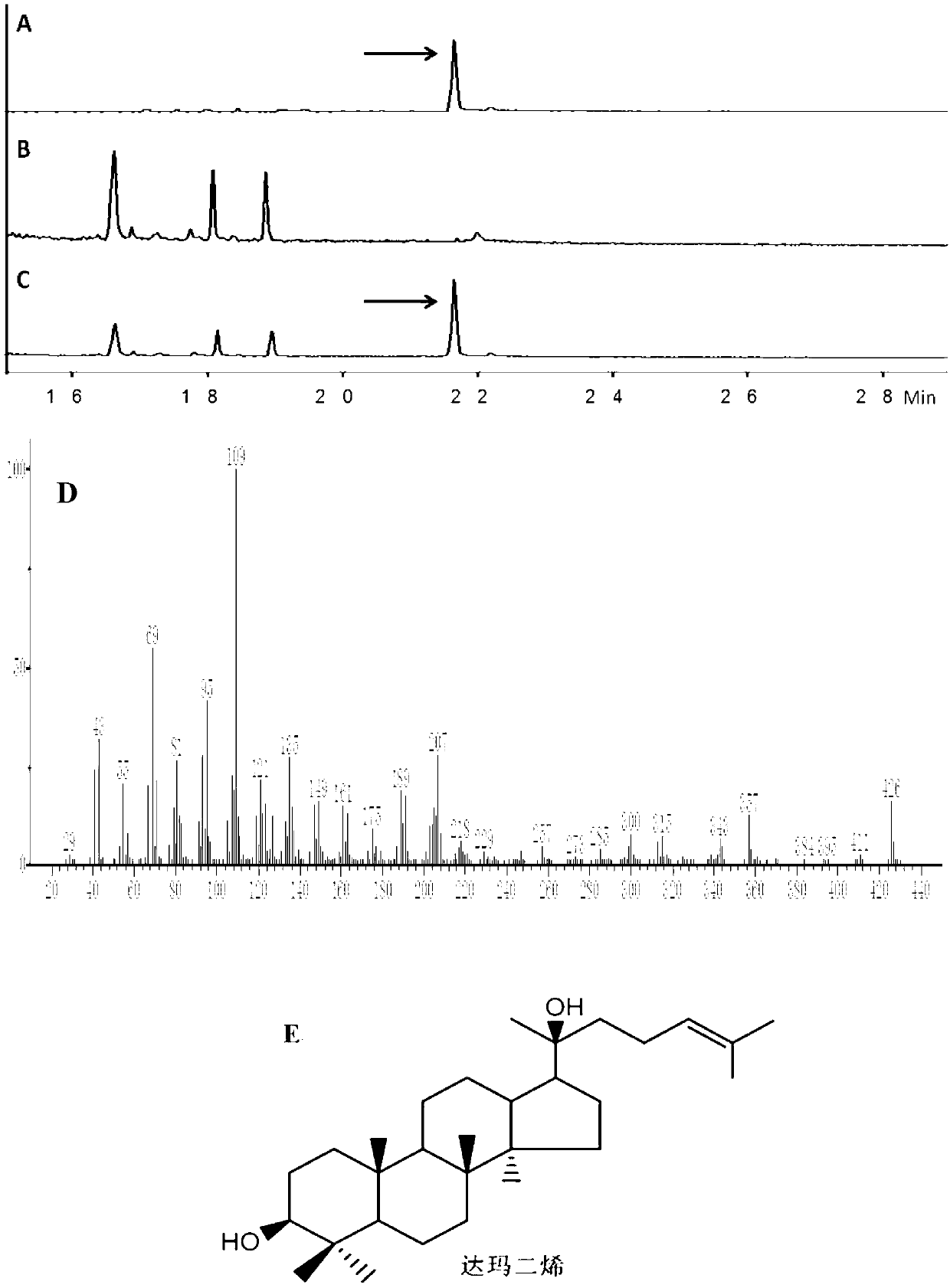
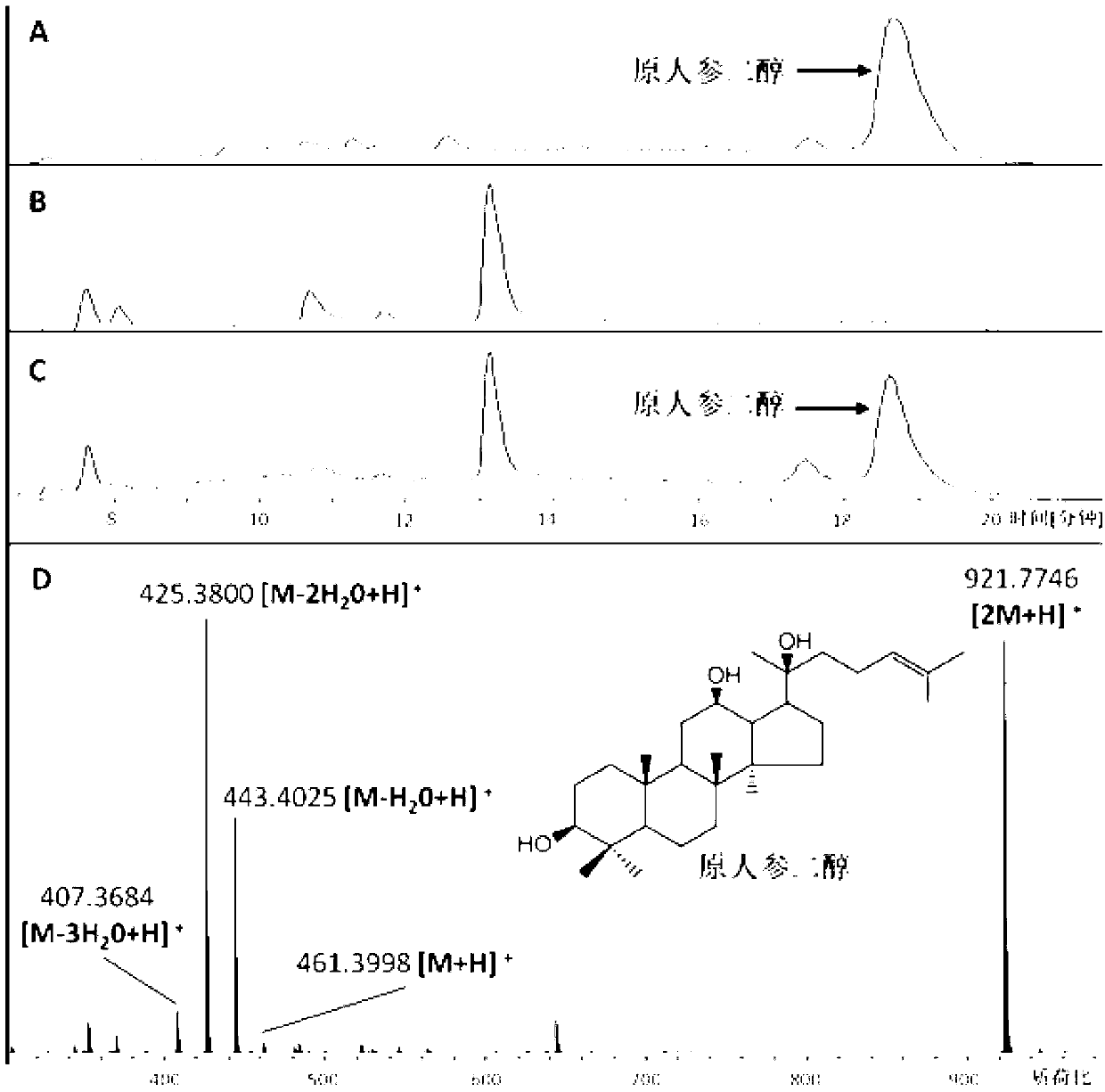
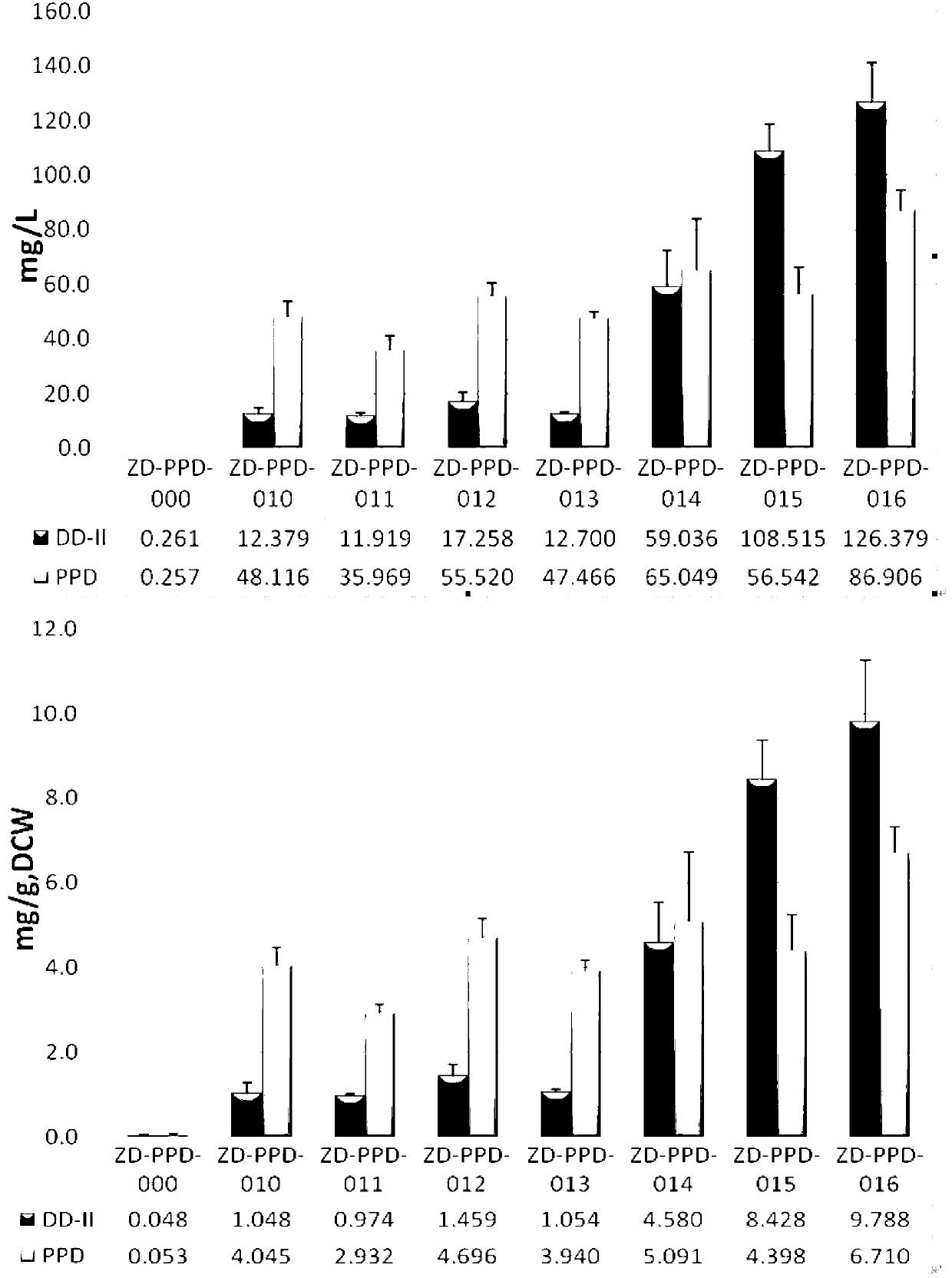
Recombinant microorganism for preparing dharma diene and protopanoxadiol and construction method thereof
A technology encoding protopanaxadiol and dammar diene synthase, which is applied in the biological field and can solve problems such as species degradation, ginsenoside compound production that cannot meet social needs, and growth environment damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Embodiment 1, gene element cloning and plasmid construction containing corresponding gene element
[0088] 1. The cloning of genetic elements is divided into the following three steps:
[0089] (1) Yeast genomic DNA extraction
[0090] Pick Saccharomyces cerevisiae BY4742 (Saccharomyces cerevisiae BY4742, recorded in Carrie baker brachmann et al., 1998, YEAST, 14:115-132, the public can obtain from Tianjin Institute of Industrial Biotechnology and Institute of Traditional Chinese Medicine, China Academy of Chinese Medical Sciences.) Plaques in YPD liquid medium (recipe: 1% Yeast Extract (yeast extract), 2% Peptone (peptone), 2% Dextrose (glucose)), 30°C, 200rpm, cultured for 24h. 10000g, 5 minutes to collect the bacteria in a 1.5ml centrifuge tube, wash twice with water, resuspend the bacteria in the yeast wall breaking solution (25ul yeast wall breaking enzyme, 470ul sorbitol buffer, 5ul β-ME), incubate at 30°C Centrifuge after 1h; resuspend the bacteria in 500ul TEN...
Embodiment 2
[0180] Construction of Saccharomyces cerevisiae Genetic Engineering Strain ZD-PPD-000
[0181] 1) rDNA-LEU2-up, P PGK1 -PgDDS-T ADH1t , P TDH3 -AtCPR1-T TPI1 , P TEF1 -PgPPDS-T CYC1 and rDNA-down preparation
[0182]Use the PCR templates and primers described in Table 14 to perform PCR to obtain functional modules: M1 (rDNA-LEU2-up), M2 (P PGK1 -PgDDS-T ADH1t ), M3 (P TDH3 -AtCPR1-T TPI1 ), M4 (P TEF1 -PgPPDS-T CYC1 ), M5 (rDNA-down) and other functional modules. The amplification system is: NewEngland Biolabs Phusion 5Xbuffer 10ul, dNTP (10mMeach dNTP) 1ul, DNA template 20ng, primers (10uM) 1ul, PhusionHigh-Fidelity DNAPolymerase (2.5U / ul) 0.5ul, add distilled water to a total volume of 50ul . The amplification conditions are 98°C pre-denaturation for 1.5 minutes (1 cycle); 98°C denaturation for 10 seconds, annealing for 10 seconds (annealing temperature 58°C), 72°C extension for 2 minutes (32 cycles); 72°C extension for 8 minutes (1 cycle), the product is recov...
Embodiment 3
[0192] Embodiment 3, the construction of Saccharomyces cerevisiae genetically engineered bacteria ZD-PPD-010
[0193] The plasmid p δ-tHMG1 was single-digested with Xho1, and the target fragment δ-tHMG (containing the expression cassette P PGK1 -tHMG1-T ADH1t ).
[0194] The same method as in Example 2 was used to prepare and transform ZD-PPD-000 competent cells, transfer into δ-tHMG1, and culture in screening culture to obtain transformants. The medium for screening culture is: 0.8% yeast selection medium SD-Ura-Trp-Leu-HIS3, 2% glucose, 0.005% HIS3., 0.01% Trp.; the conditions for screening culture are: 30 degrees, culture for more than 36 hours.
[0195] Using Sac11-pGK1 and Sac11-Pme-ADHt primers for PCR identification, the fragment of about 2539bp was the correct positive clone, named strain ZD-PPD-010.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com