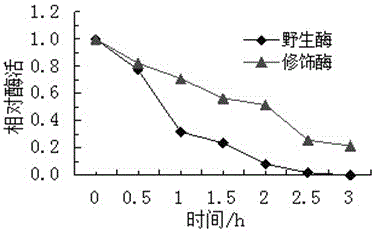
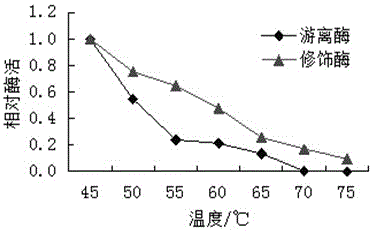
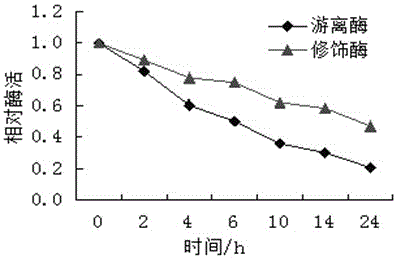
A method for modifying oxalate decarboxylase with monomethoxypolyethylene glycol
An oxalate decarboxylase, polyethylene glycol technology, applied in microorganism-based methods, biochemical equipment and methods, enzymes, etc., can solve problems such as denaturation and inactivation, and achieve low cost, enhanced tolerance, and mild conditions. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] (1) Preparation of oxalate decarboxylase
[0023] Ferment and cultivate the genetically engineered bacteria E.coliBL21(DE3) / pET32a / YvrK, add 0.4mmol / LIPTG to induce the expression of oxalate decarboxylase, harvest the bacteria and extract with 50mmol / LpH8.0 phosphate buffer to obtain a crude enzyme solution, use fast protein solution Phase chromatography system AKTA-FPLC was used for enzyme purification.
[0024] (2) Activation of mPEG
[0025] Weigh 1g of mPEG and dissolve it in 1.5mL of chloroform, add acetic anhydride at the ratio of acetic anhydride / mPEG molar ratio of 20:1, add 6mL of DMSO dried by calcium hydride at the same time, stir and react at room temperature for 9h, then add the mixed solution dropwise to 4 Double the volume of diethyl ether in an ice bath and stir at high speed. The precipitate was dissolved in 2 mL of chloroform, and the precipitation was repeated three times. The precipitate was vacuum-dried at room temperature for 24 h, and stored for...
Embodiment 2
[0037] (1) Preparation of oxalate decarboxylase
[0038] Same as (1) operation in the embodiment
[0039] (2) Activation of mPEG
[0040] Operation with (2) in Example 1.
[0041] (3) mPEG modified oxalate decarboxylase
[0042] Put mPEG5000-aldehyde and Oxdc enzyme protein at a molar ratio of 10:1 to 150:1 at 37°C and pH 5.0 and stir for 12 hours, then add 20-fold molar ratio of glycine to the enzyme protein to terminate the reaction. The reaction solution was put into a 8000-14000 dialysis bag, and dialyzed in PEG20000 for 12 hours. The final solution was centrifuged. The modification rate and enzyme activity recovery rate were determined by TNBS method as shown in the table below.
[0043] Aldehyde / enzyme (molar ratio) 10:1 20:1 30:1 40:1 50:1 100:1 150:1 Modification rate (%) 7.54 11.96 18.17 25.30 39.12 25.13 10.25 Enzyme Recovery 97.67 94.57 87.60 83.72 78.29 40.31 12.40
[0044] When the mPEG / enzyme molar ratio is lower...
Embodiment 3
[0046] (1) Preparation of oxalate decarboxylase
[0047] Same as (1) operation in the embodiment
[0048] (2) Activation of mPEG
[0049] Operation with (2) in Example 1.
[0050] (3) mPEG modified oxalate decarboxylase
[0051] Put mPEG5000-aldehyde and Oxdc enzyme protein at a molar ratio of 50:1 at 4-45°C and pH 5.0 and stir for 12 hours, then add 20-fold molar ratio of glycine to the enzyme protein to terminate the reaction. The reaction solution was put into a 8000-14000 dialysis bag, and dialyzed in PEG20000 for 12 hours. The final solution was centrifuged. The modification rate and enzyme activity recovery rate were determined by TNBS method as shown in the table below.
[0052] temperature °C 4 25 37 45 Modification rate (%) 14.88 24.31 39.02 56.98 Enzyme Recovery 92.71 89.77 67.21 35.12
[0053] The effect of mPEG-modified oxalate decarboxylase was compared at different temperatures, and the results are shown in the table abov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com