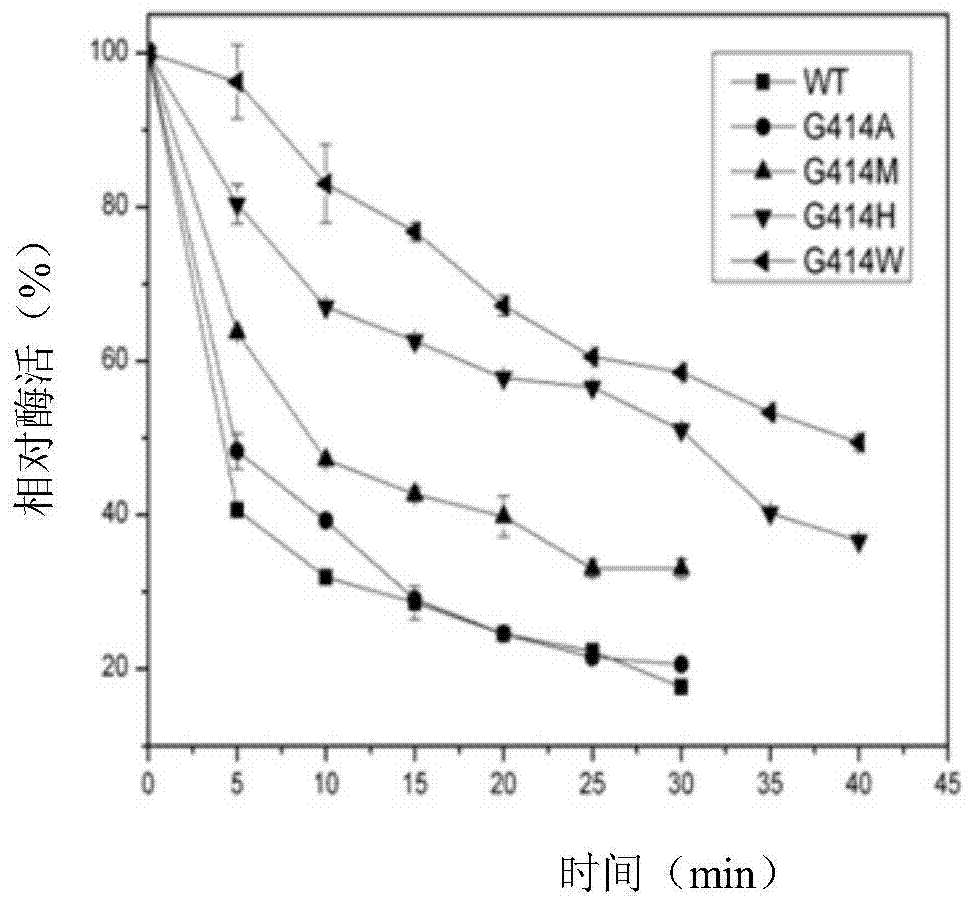
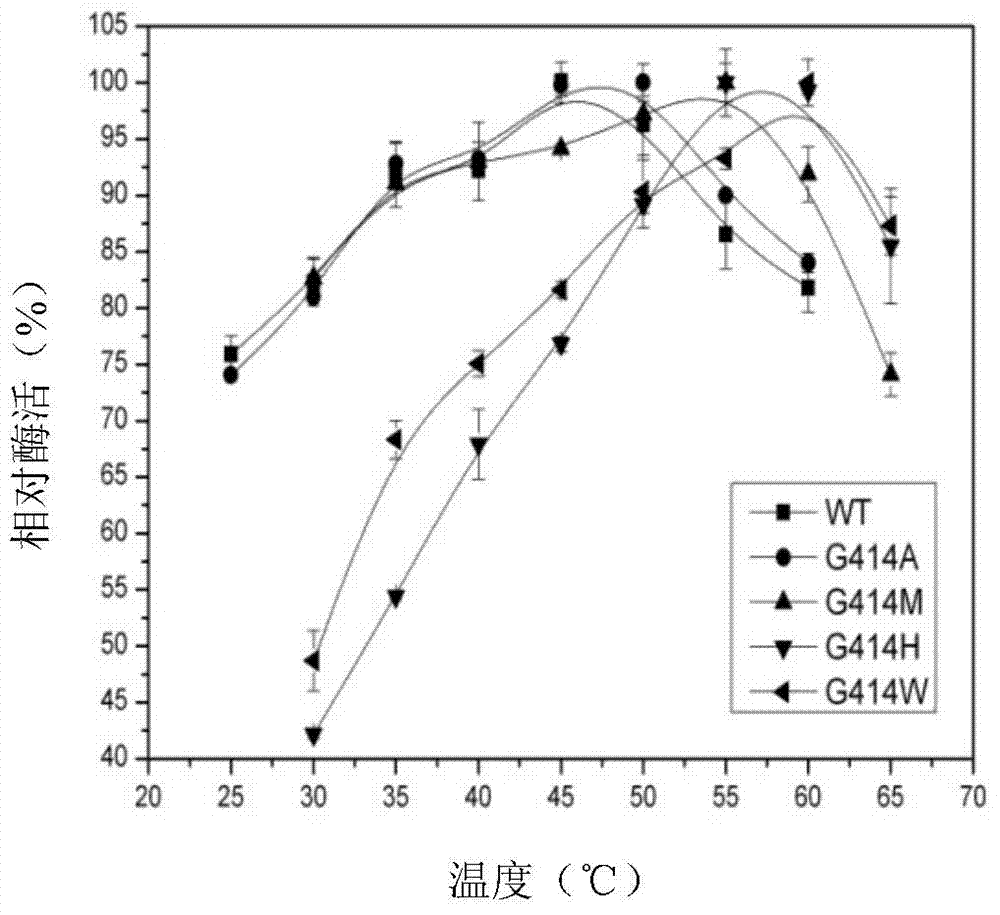
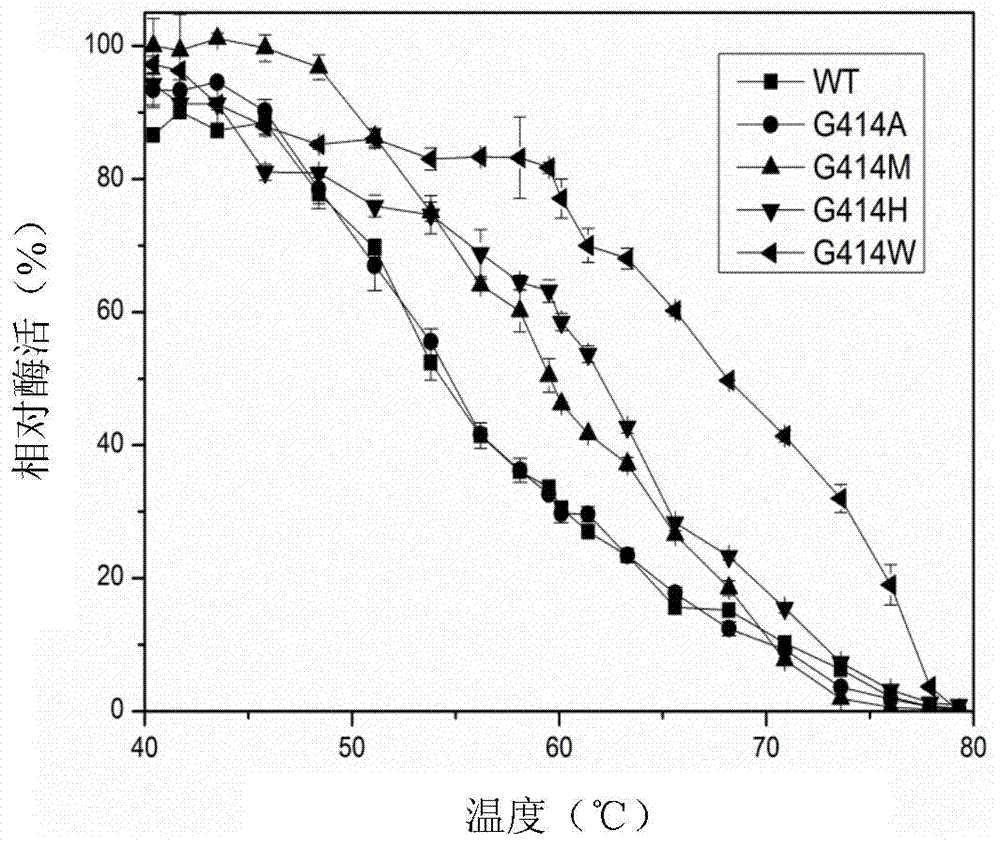
Candida rugosa lipase 1 mutant and gene
A technology of Candida folds and lipase, applied in the field of protein engineering, can solve problems such as unreported, little thermal stability, etc., and achieve the effect of improving application potential and improving thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Cloning of Example 1 Wild-type Candida rugosa Lipase 1
[0023] Candida plicata lipase 1 gene was amplified by designing upstream and downstream primers, upstream primers:
[0024] 5'-TCT CTCGAG AAAAAGACATCATCACCACCATCATGCCCCCACCGCCACGCT-3'; the underlined part is the Xho Ⅰ site; downstream primers:
[0025] 5'-CGG TCTAGA TAACACAAAGAAAGACGGCGGGTTGGAGA-3', the underlined part is the Xba I site.
[0026] Using LA Taq polymerase (TaKaRa Company), the target gene LIP1 was amplified with the participation of upstream and downstream primers; PCR reaction conditions were: 95°C pre-denaturation for 10 min, each cycle including 95°C denaturation for 25s, 60°C annealing for 30s, 72 Extend at ℃ for 90s, a total of 30 cycles; finally derivatize at 72℃ for 5min. After the reaction, the PCR products were detected on a 1% agarose gel. The length of the nucleic acid is consistent with the size published in the PDB database (1602bp), and its nucleotide sequence is shown as SEQ ID...
Embodiment 2
[0028] Example 2 Construction of gene mutation library by site-directed saturation mutation
[0029] Primers for site-directed saturation mutagenesis by:
[0030] Upstream primer, 5'-CTCGGCGACCTT NNK TTTACGCTTGCTCGTCGCTAC-3';
[0031] Downstream primer, 5'-CTCGGCGACCTT MNN TTTACGCTTGCTCGTCGCTAC-3',
[0032]The underlined part is the saturation mutation site. Using PrimerSTAR max DNA polymerase (TaKaRa Company), the whole plasmid was amplified with the participation of upstream and downstream saturation mutation primers. PCR reaction conditions: pre-denaturation at 98°C for 5 min, each cycle of denaturation at 98°C for 10 s, annealing at 55°C for 5 s, extension at 72°C for 2.5 min, a total of 30 cycles; the final extension at 72°C for 5 min. After the PCR product was purified, it was directly transformed into Escherichia coli DH5α competent, heat-shocked at 42°C for 90s, and incubated at 37°C for 1h. Then spread it on the LB plate containing 25μg / ml Zeocin resistance. A...
Embodiment 3
[0034] Example 3 Expression and purification of Candida rugosa lipase 1 wild type and mutant
[0035] Inoculate Pichia pastoris with integrated LIP1 gene into 4mL YPD medium containing 25μg / mL Zeocin, culture at 30°C for 2 days, then inoculate into 200mL fresh medium of YPD, culture at 30°C, 220rpm for 72h, and harvest the cells . Centrifuge at 8000 rpm for 30 min to harvest the supernatant enzyme solution, and filter the crude enzyme solution with a 0.22 μm filter membrane to remove impurities in the crude enzyme solution. Afterwards, use an ultrafiltration membrane device with a molecular weight cut-off of 10,000 to concentrate and decolorize the sample. During the ultrafiltration process, add 50mM Tris-HCL pH7.5 buffer to dilute the enzyme solution until the ultrafiltration membrane discharge is colorless can be stopped.
[0036] Since the N-terminus of the expressed protein contains a histidine tag, the concentrated sample can be purified by a nickel affinity column. Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com