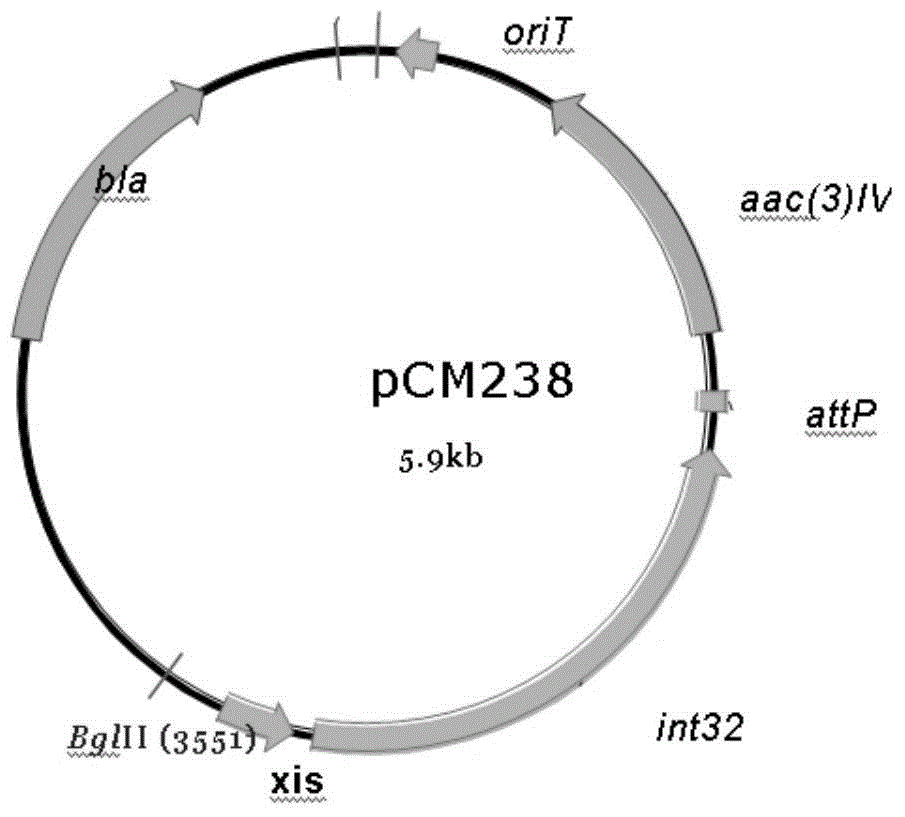
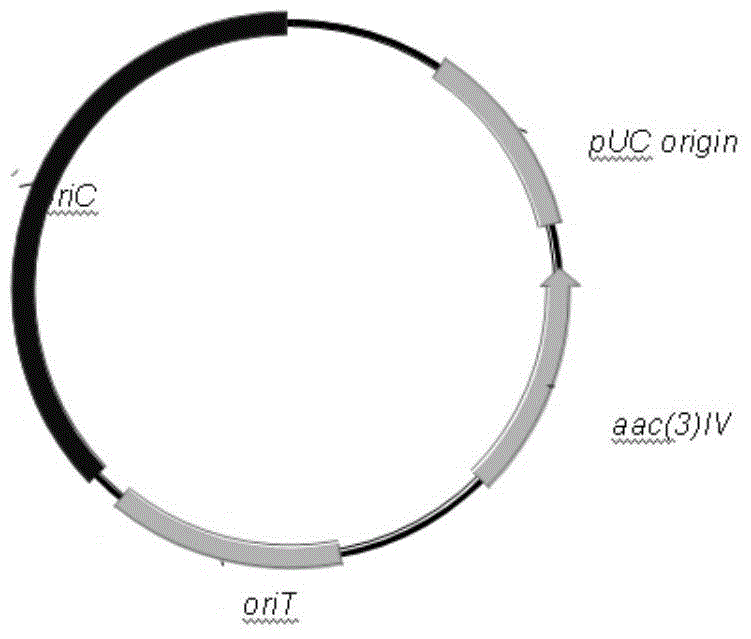
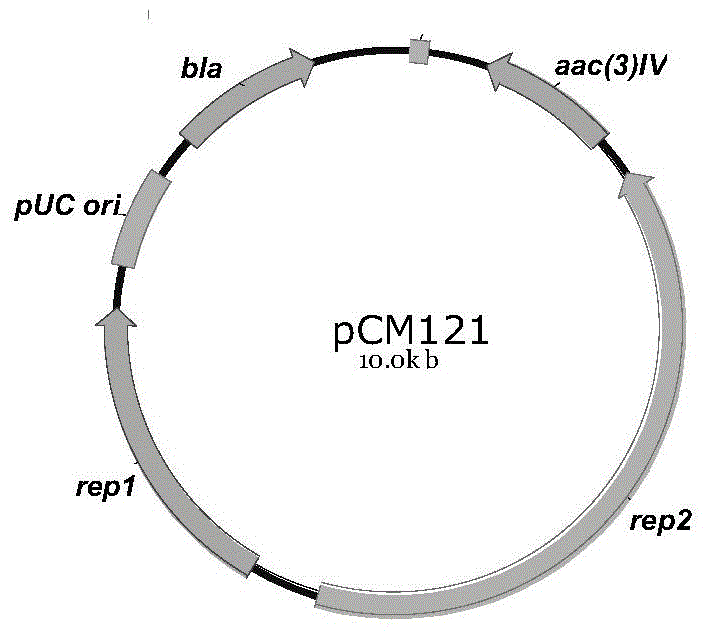
Novel integrase and application thereof to efficiently modifying saccharopolyspora spinosa
A technology of Saccharopolyspora spinosa and integrase, applied in applications, enzymes, fungi, etc., can solve problems such as lack of attB, unstable genetics, and difficult genetic operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0191] Embodiment 1: Introduction and stability test of plasmid
[0192] The constructed plasmids were respectively transformed into Escherichia coli S17-1, and transformed into S. spinosa by E. coli-Saccharopolyspora indirect conjugative transfer. The purified resistant colonies were uploaded on the non-resistant plate for the first and second generations, and the spores were collected and diluted to obtain a single colony. Pick 100-200 single colonies on the resistant plate and non-resistant plate respectively, culture at 30°C, and detect the proportion of non-resistant colonies (colonies with plasmid loss). The results are shown in Table 3.
[0193] Table 3 Plasmid Stability Test
[0194]
[0195]
[0196] The results show that the stability of autonomously replicating plasmids in S. spinosa is different. After non-resistance selection, the loss rate of plasmids is high, which can be used to selectively lose plasmids in the shuffling screening of circulating genomes...
Embodiment 2
[0197] Example 2: Spinosyn production by fermentation of Saccharopolyspora spinosa.
[0198] The Saccharopolyspora spinosa spores on the mature slant were inoculated in the seed shaker flask with a sterile inoculation shovel, the amount of the seed shaker flask was 25ml / 250ml, and cultivated at 28°C and 220r / min for 64-72h. Insert the fermentation medium with 10% inoculum amount, the volume of the fermentation shaker flask is 25ml / 250ml shaker flask, cultivate at 28°C 220r / min for 8-10d,
[0199] Sampling was performed to detect spinosyn production.
[0200] HPLC analysis conditions for determination of spinosyn yield: chromatographic column: Agilent Zorbax Eclipse XDB-C8 4.6×12.5 (5 μm); detection wavelength: 246nm; mobile phase: methanol: acetonitrile: water = 45:45:10, plus 0.005% ammonium acetate ; Flow rate: 1.0ml / min.
[0201] The formula of Saccharopolyspora spinosa slant medium is: whole milk powder 2%, glucose 0.5%, yeast extract powder 0.3%
[0202] The formula of...
Embodiment 3
[0204] Example 3: Application of resistance plasmid markers in genome shuffling screening of Saccharopolyspora spinosa
[0205] The bacterial strain SN304 containing the hygromycin resistance gene plasmid pSN11 marker and the bacterial strain SN0207 containing the apramycin resistance gene plasmid pWT295 marker were respectively prepared into protoplasts, and the protoplast fusion test was carried out according to the principle of patent ZL200710043687.7 ( Refer to literature 12 for the specific method, randomly pick 100 fusion sons for fermentation detection, and the fermentation titer increases by 39%, and the highest increase reaches 41% ( Figure 4 ). The fusion son can further lose one of the resistances, combined with physical or chemical mutagenesis, to obtain a compound parent, and perform the next round of genome shuffling, and repeat multiple rounds of fusion to screen for high-yielding strains.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com